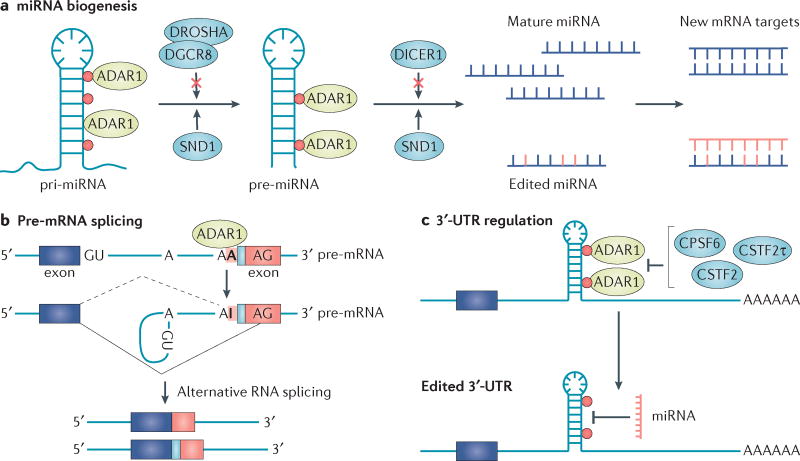
Figure 2. Consequences of RNA editing by ADAR.
a | Adenosine-to-inosine (A-to-I) editing-dependent regulation of microRNA (miRNA) biogenesis and targeting occurs at multiple stages. Editing (depicted by red dots) of primary miRNAs (pri-miRNAs) by adenosine deaminase acting on double-stranded RNA 1 (ADAR1) and ADAR2 prevents processing by the microprocessor complex composed of DROSHA and DGCR8, which results in decreased production of mature miRNA. Similarly, RNA editing of precursor miRNA (pre-miRNA) affects the DICER1 cleavage process. Both edited pri-miRNAs and edited pre-miRNAs are specifically recognized and degraded by the ribonuclease staphylococcal nuclease domain-containing 1 (SND1). Finally, editing within the seed region of mature miRNAs potentially changes the specific mRNA target compared with the unedited version. b | RNA editing at alternative 3′-acceptor sites converts the intronic AA into AI dinucleotides, which mimic the conserved AG sequences normally found at 3′-splicing sites. This introduction of a new splicing acceptor site results in an alternatively spliced mRNA with an insertion (light blue). The dashed line represents an alternative splicing event generated by A-to-I editing. c | ADAR1 competes with canonical 3′-untranslated region (UTR) processing factors (cleavage stimulation factor subunit 2 (CSTF2), CSTF2τ and cleavage and polyadenylation-specific factor 6 (CPSF6)) by direct binding to 3′-UTRs shortly after they are transcribed. As miRNAs predominately target mRNA 3′-UTR sites, RNA editing changes within the miRNA target sequences may prevent miRNA binding and therefore downregulation of the mRNA.