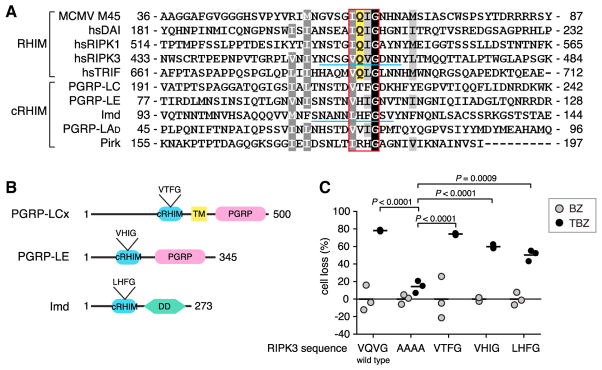
Figure 1. Conserved Function of Cryptic RHIM Motifs in the DrosophilaImd Pathway.
(A) cRHIMs of D. melanogaster proteins aligned with human and MCMV RHIM-sequences. The four core amino acids are boxed with red. The amyloidogenic Q that is highly conserved in mammalian sequences but missing in Drosophila cRHIM is highlighted with yellow. Shading represents conservation according to Blosum 62 scoring matrix. Region underlined with blue represents the sequence in RIPK3 that was replaced with the underlined sequence from Imd in Figure 6H.
(B) Schematic illustration of PGRP-LCx, PGRP-LE, and Imd domain composition. Abbreviations are as follows: TM, transmembrane domain; PGRP, PGRP domain; DD, death domain.
(C) Effects of RIPK3 substitution mutants on TNF-induced necroptosis in HeLa cells. Cells were transfected with wild-type or chimeric RIPK3 where the VQVG core motif was substituted with AAAA, VTFG (PGRP-LC), VHIG (PGRP-LE), or LHFG (Imd), and necroptosis activated with combined TNF (T), BV6 (B), and zVAD-fmk (Z) treatment. The graph shows individual data points (N = 3) with mean indicated as a line. BZ indicates no TNF stimulation (gray dots); TBZ indicates BV6 and zVAD-fmk-treated cells stimulated with TNFα (black dots).
See also alignment of selected insect cRHIMs in Figure S1.