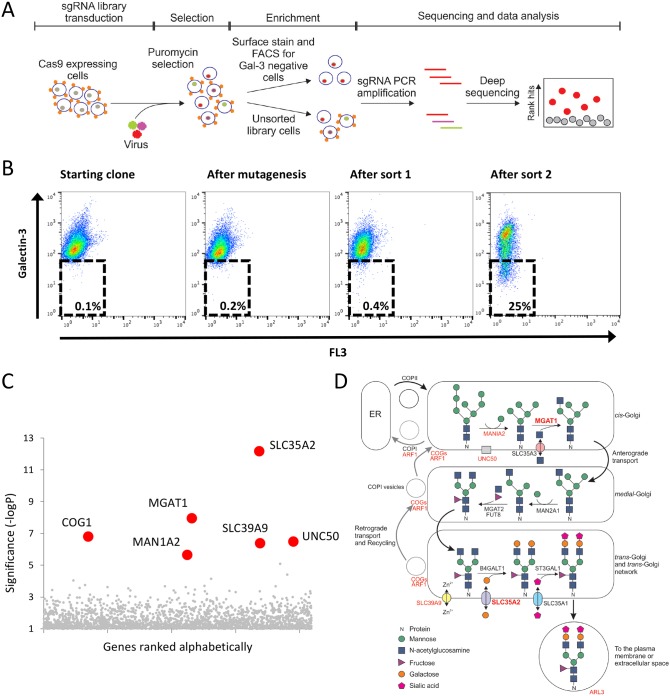
Fig. 1.
A CRISPR/Cas9-mediated genetic screen identifies genes required for cell surface localization of Gal-3. (A) Schematic of the CRISPR/Cas9 screen in sHeLa to identify genes required for Gal-3 cell surface localization. Cells were transduced with a lentiviral sgRNA library (sgRNA transduction indicated by colours in the nucleus) and cells that were successfully transduced were selected with puromycin. After selection, the population was split into two; one half was sorted by FACS to enrich for cells that have less Gal-3 on the surface (Gal-3 is represented by small orange shapes on the cell surface) and the other was not sorted to represent the entire library. After two rounds of enrichment, the DNA from both the enriched population and the unsorted library was harvested, and enriched sgRNAs were identified by sequencing. Targeted genes were then plotted according to their relative enrichment. (B) CRISPR-mediated mutagenesis was performed on sHeLa using the GeCKO v2 sgRNA library, and rare cells with decreased surface Gal-3 expression were selected by two sequential rounds of FACS. Cell surface Gal-3 was measured on live cells using an anti-Gal-3 antibody conjugated to Alexa Fluor 647. (C) Plot illustrating the hits from the CRISPR screen. The RSA algorithm was used to identify the significantly enriched genes targeted in the selected cells. The most significantly enriched genes are labelled. (D) Schematic of the N-linked glycosylation pathway within the Golgi. Genes identified to be important for Gal-3 surface localization by the CRISPR screen are highlighted in red, and those chosen for further study (MGAT1 and SLC35A2) are shown in bold.