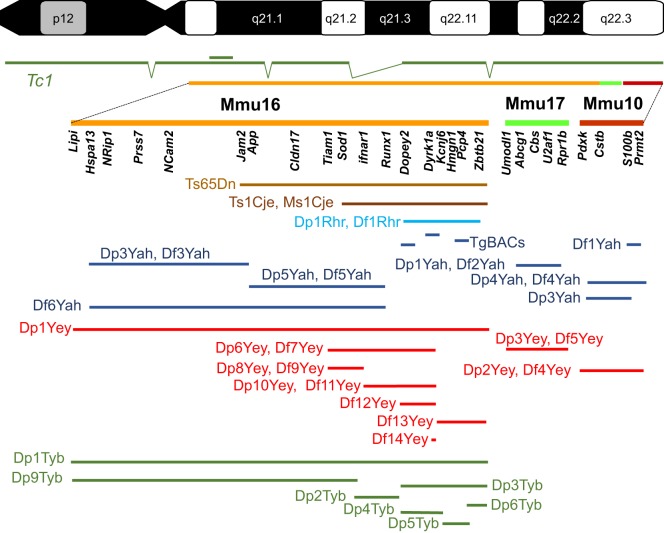
Fig. 1.
Mouse models of DS. Human chromosome 21 (p and q arms; G-banding) is depicted at the top of the figure, with the mouse genome orthologous region found on chromosome 16 (Mmu16), Mmu10 and Mmu17 shown respectively in orange, light green and red. A few known genes that are homologous to Hsa21 genes in the DS critical region are listed below each chromosome. The transchromosomic Tc1 mouse model is shown in dark green, with deletions and a duplication (double bar) relative to Hsa21 depicted. Below, the segment of the DS critical region encompassed in different mouse models for DS is illustrated. The original Ts65Dn (Reeves et al., 1995) and Ts1Cje (Sago et al., 1998) models (shown in brown) originated by accidental translocation of Mmu16 segments respectively on Mmu17 and Mmu12, with some additional changes (Duchon et al., 2011b; Reinholdt et al., 2011). Olson et al. (2004) published the first engineered duplication (Dp) and deletion [deficiency (Df)] for the DS critical region (light blue). New models have been developed in the last 10 years by the authors of this Review, as shown in dark blue (Duchon et al., 2011a; Lopes Pereira et al., 2009; Besson et al., 2007; Marechal et al., 2015; Sahun et al., 2014; Raveau et al., 2012; Arbogast et al., 2015; Brault et al., 2015b), red (Jiang et al., 2015; Liu et al., 2011, 2014; Yu et al., 2010a,b,c; Li et al., 2007) and green (Lana-Elola et al., 2016). TgBACs, a few models for BAC or PAC (P1-derived artificial chromosome) transgenic lines.