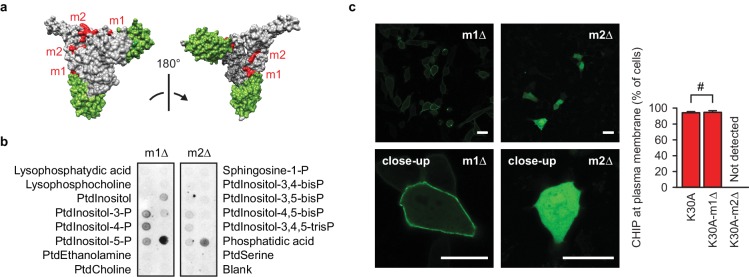
Figure 3. A positively charged patch is required for CHIP binding to phospholipids.
(a) Surface representation of CHIP dimer (PDB entry 2c2l) with TPR domains colored green. m1, m2, positively charged patches colored red. (b) m2-mutation (m2Δ) affects CHIP interaction with phospholipids in vitro as determined by lipid-binding assay. One representative out of three independent experiments is shown. (c) m2Δ CHIP loses association with membranes in vivo. Scale bar 20 μm. #, no statistically significant difference according to chi-square analysis; N = 3 independent experiments (mean ± SD).