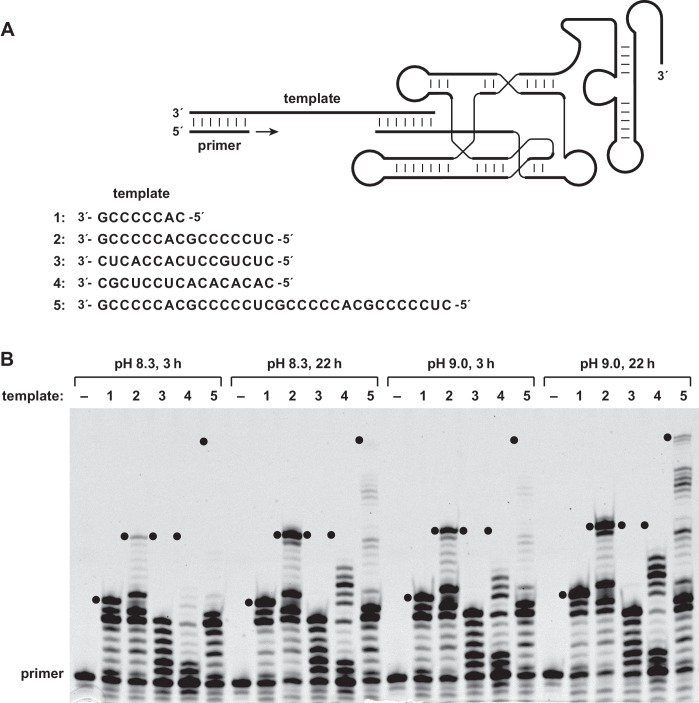
Figure 1. Reverse transcriptase activity of the 24–3 ribozyme.
(A) Secondary structure of the complex formed by the ribozyme, template, and primer (nucleotide sequences are listed in supplementary file 1). The template consists of four regions: primer binding site, sequence to be copied, A3 or A5 spacer, and ribozyme-pairing domain (listed 3´→5´). The ribozyme was tested for its ability to copy five different template sequences (1–5). For sequences of other regions of the template, see supplementary file 1. (B) Extension of a deoxynucleotide-terminated RNA primer on an RNA template. Reaction conditions: 100 nM ribozyme, 125 nM template, 125 nM primer, 2 mM each dNTP, 200 mM MgCl2, pH 8.3 or 9.0, 20°C, 3 or 22 hr. Black dots indicate the expected position of full-length products.
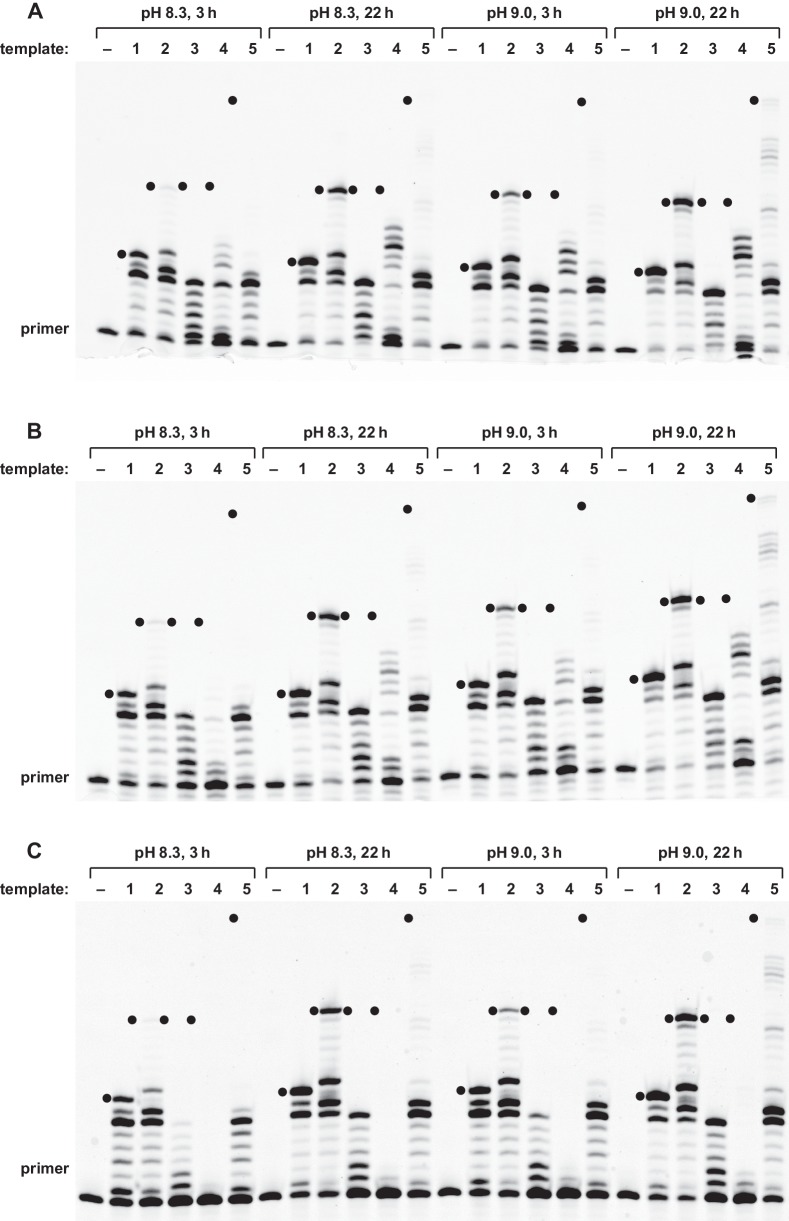
Figure 1—figure supplement 1. Reverse transcriptase activity of the 24–3 ribozyme.
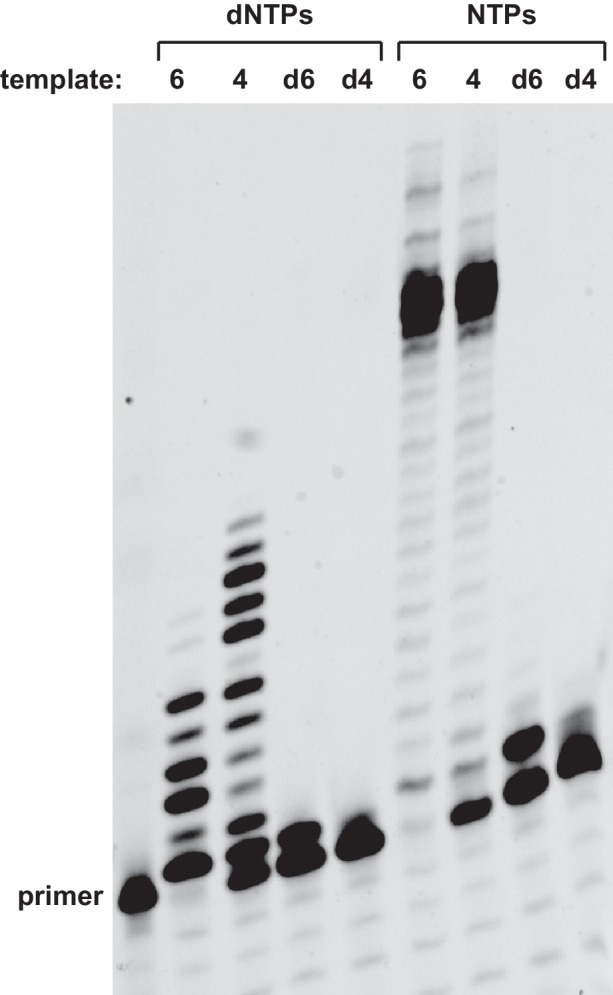
Figure 1—figure supplement 2. Lack of DNA-dependent polymerase activity of the 24–3 ribozyme.