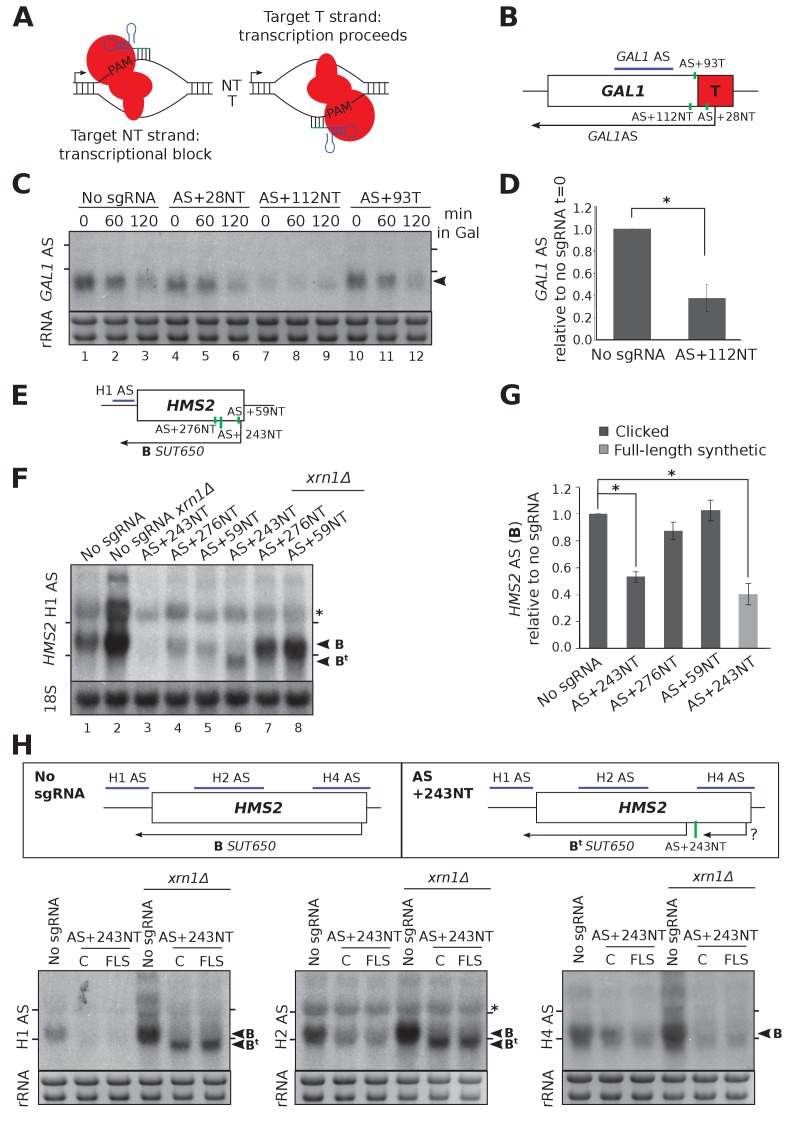
Figure 2. CRISPRi reduces antisense transcripts at GAL1 and HMS2.
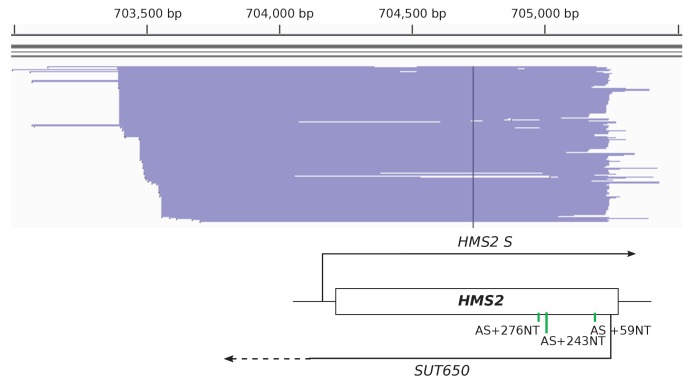
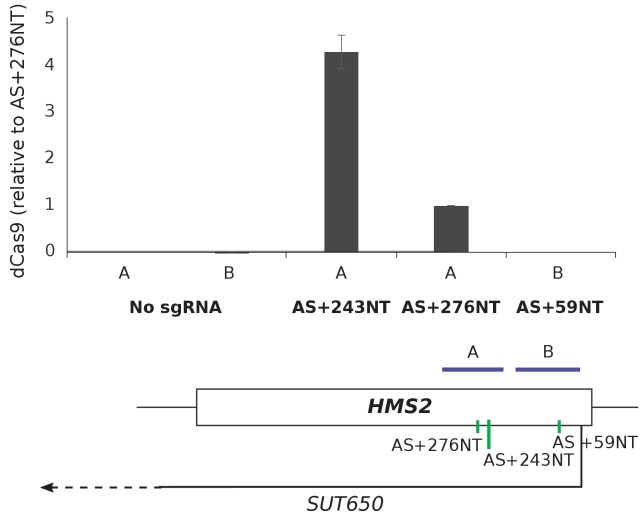
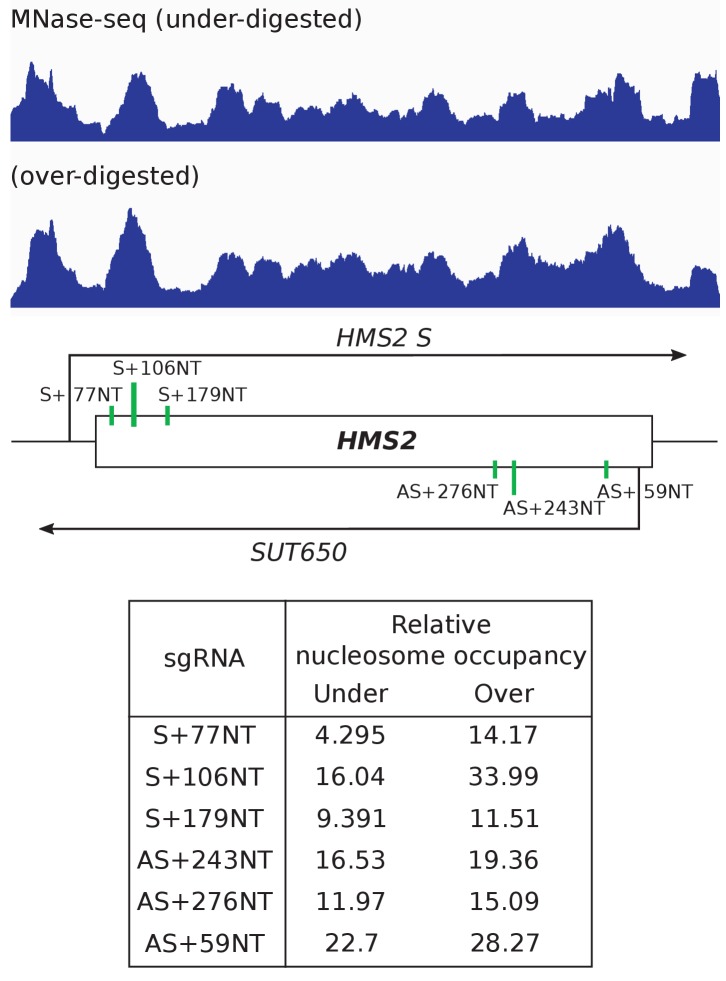
(A) A schematic demonstrating the previously reported requirement of non-template strand targeting by the sgRNA/dCas9 complex for strand-specific transcriptional repression. Arrows indicate the direction of transcription. NT, non-template; T, template; red, dCas9; PAM, protospacer adjacent motif; green/blue, sgRNA. (B) A schematic of the engineered GAL1 locus showing the site of insertion of the ADH1 terminator (T, red box), the transcription start site for the stable GAL1 antisense transcript and the positions of the designed sgRNAs (green vertical lines) targeting the non-template (AS+28NT, AS+112NT) and template (AS+93NT) strands with respect to antisense transcription. The position of the Northern blotting probe detecting the GAL1 AS transcript is shown in purple. (C) A Northern blot showing the reduction in GAL1 antisense transcript (black arrowhead) in the strain expressing sgRNA AS+112NT/dCas9 relative to the no sgRNA control (GAL1:ADH1t snR52::URA3 with dCas9). sgRNAs AS+28NT and AS+93T do not alter GAL1 AS levels. Samples were taken from cells grown in glucose (t = 0) and at the indicated times after transfer to galactose-containing media (min). Positions of the rRNA are indicated by the short horizontal lines. Ethidium-bromide-stained rRNA is used as loading control. (D) Quantification of Northern blotting for the GAL1 AS transcript at t = 0 in the control no sgRNA strain and the strain with sgRNA AS+112NT. N = 6, errors are SEM, *p=0.004. See Source data 1, tabs 10–12. (E) A map of the HMS2 locus showing the HMS2 AS transcript, SUT650 (black arrow, transcript B) and positions of the three sgRNAs targeting SUT650 (green vertical lines). The position of the Northern blotting probe to detect SUT650 (H1 AS) is shown by the purple line. (F) A Northern blot probed with HMS2 antisense probe H1 (see schematic in (E)) showing the level of SUT650 (black arrowhead, transcript B) in the no sgRNA control (snR52::URA3 with dCas9) strain and strains expressing the indicated antisense-targeting sgRNAs. Deletion of XRN1 allows detection of a shorter antisense transcript (Bt) in the strain expressing AS+243NT. Positions of the rRNA are indicated by the short horizontal lines. *Represents cross-hybridisation with the 25S rRNA. A blot probed for the 18S rRNA is used as loading control. (G) Quantification of the level of SUT650 (transcript B) reduction in the strains expressing each of the three antisense sgRNAs relative to the control no sgRNA strain. N = 4–8, errors represent SEM, *p<0.05. Click-linked and full-length synthetic sgRNA AS+243NT templates behave similarly. See Source data 1, tabs 5–9. (H) Northern blots with a series of HMS2 antisense-specific probes. A new shorter antisense transcript (Bt) can be detected upon XRN1 deletion in the strains expressing sgRNA AS+243NT/dCas9. Two clones of the CRISPRi strains produced using clicked (C) or full-length synthetic (FLS) DNA oligos for strain construction are shown. Positions of the antisense-specific probes (purple) and the site of sgRNA AS+243NT/dCas9 binding (green vertical line) are shown in the schematic. Positions of the rRNA on the Northern blot are indicated by the short horizontal lines. *Represents cross-hybridisation with the 25S rRNA. Ethidium-bromide-stained rRNA is used as loading control.