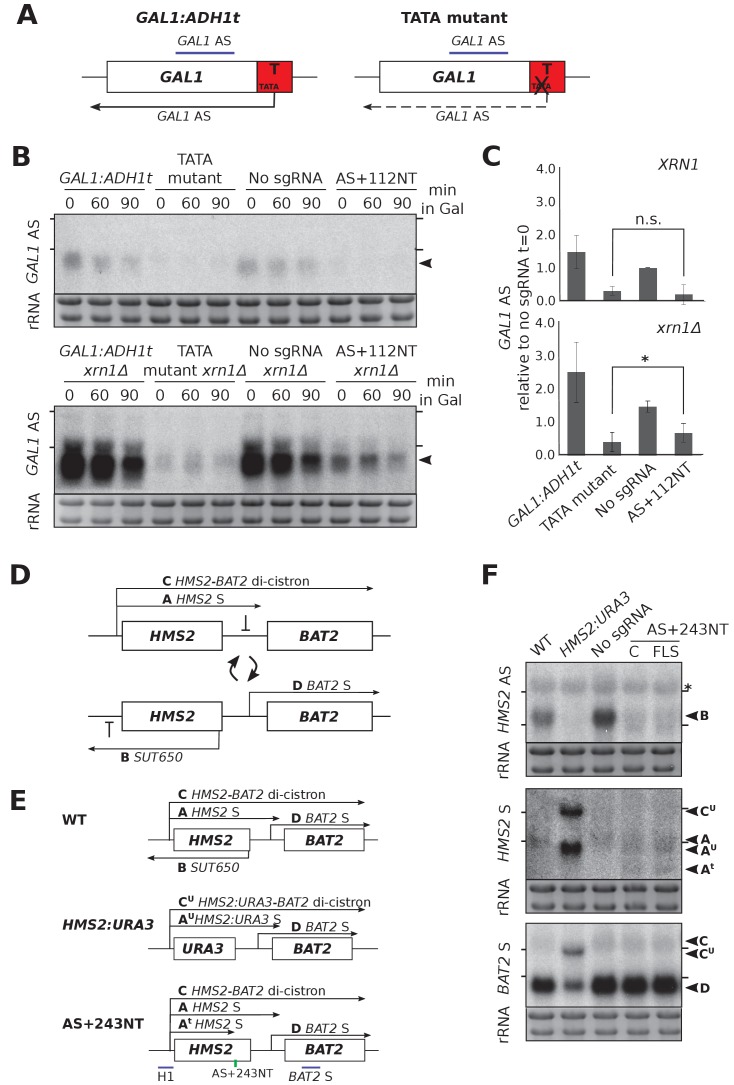
Figure 4. CRISPRi block of SUT650 and GAL1 AS is not as efficient as previous genetic interventions.
(A) A schematic showing the previously used genetic intervention to repress the GAL1 AS transcript (Murray et al., 2015). In the TATA mutant construct, a TATA box within the engineered ADH1 terminator (T) is scrambled and the level of GAL1 antisense transcript is reduced. (B) A Northern blot probed for the GAL1 antisense transcript (black arrowhead) comparing the two methods of reducing antisense at this locus in the presence (top blot) or absence (bottom blot) of XRN1. All samples were run on the same gel and the blots were exposed to film for the same time. Samples were taken from cells grown in glucose (t = 0) and at the indicated times after transfer to galactose-containing media (min). Positions of the rRNA are indicated by the short horizontal lines. Ethidium-bromide-stained rRNA is used as loading control. (C) Quantification of the Northern blot in (B) at t = 0. N = 2, errors represent the SEM, *p=0.026, n.s. non-significant. See Source data 1, tab 12. (D) A schematic of the HMS2-BAT2 locus, showing the HMS2 sense and antisense transcripts (A and B), the HMS2-BAT2 di-cistronic transcripts (C and C’) and the BAT2 sense transcript (D). Cells switch between sense- and antisense-dominant states in which the indicated transcripts are produced (Nguyen et al., 2014). (E) The HMS2-BAT2 locus and associated transcripts in the wild-type (WT) strain and strains that repress the SUT650 transcript. In the HMS2:URA3 strain, the entire coding region of HMS2 is replaced by that of URA3. (F) Northern blots comparing the efficiency of SUT650 reduction by replacement of the HMS2 coding region with that of URA3 versus CRISPRi with sgRNA AS+243NT and the relative effects of these two strategies on the HMS2 sense and BAT2 transcripts shown in (E). The WT (BY4741) and HMS2.URA3 strains have been transformed with plasmid pRS315 to allow for growth on complete synthetic media lacking leucine and thus comparisons with the CRISPRi strains. Positions of the rRNA are indicated by the short horizontal lines. *Represents cross-hybridisation with the 25S rRNA. Ethidium-bromide-stained rRNA is used as loading control.