Abstract
Oxidized phospholipids (OxPLs) are widely held to be associated with various diseases, such as arteriosclerosis, diabetes, and cancer. To characterize the structure-specific behavior of OxPLs and their physiological relevance, we developed a comprehensive analytical method by establishing a measured MS/MS spectra library of OxPLs. Biogenic OxPLs were prepared by the addition of specific oxidized fatty acids to cultured cells, where they were incorporated into cellular phospholipids, and untargeted lipidomics by LC-quadrupole/TOF-MS was applied to collect MS/MS spectra for the OxPLs. Based on the measured MS/MS spectra for about 400 molecular species of the biogenic OxPLs, we developed a broad-targeted lipidomics system using triple quadrupole MS. Separation precision of structural isomers was optimized by multiple reaction monitoring analysis and this system enabled us to detect OxPLs at levels as low as 10 fmol. When applied to biological samples, i.e., mouse peritoneal macrophages, this system enabled us to monitor a series of OxPLs endogenously produced in a 12/15-lipoxygenase-dependent manner. This advanced analytical method will be useful to elucidate the structure-specific behavior of OxPLs and their physiological relevance in vivo.
Keywords: lipidomics, lipoxygenase, mass spectrometry, oxidized lipids, untargeted lipidomics, broad-targeted lipidomics, tandem mass spectrometry
Phospholipids (PLs), the main components of cell membranes, have important roles in biological systems, including in signal transduction, membrane trafficking, cell proliferation, and fusion. PLs have a glycerophosphate backbone structure, and the polar head group is linked to the sn-3 position and the acyl chains are linked to the sn-1/sn-2 positions. Saturated or mono-unsaturated fatty acids, such as palmitic acid (16:0), stearic acid (18:0), and oleic acid (18:1), are mainly located in the sn-1 position. On the other hand, PUFAs, such as linoleic acid (18:2), arachidonic acid (AA; 20:4), EPA (20:5), and DHA (22:6), are mainly located in the sn-2 position. Thus, oxidized PUFAs that are generated by oxygenases and/or by reactive oxygen species are usually located in the sn-2 position of PLs. Some of those oxidized PLs (OxPLs) are biologically active and are thought to be associated with pathogenesis of various diseases, such as arteriosclerosis, diabetes, and cancer (1–4). However, the biosynthesis and metabolism of each OxPL molecular species has not been well-characterized.
HPLC-UV and/or the analysis of thiobarbituric acid-reactive substances have conventionally been applied to monitor OxPL levels (5, 6). However, those methods are not designed to determine differences in the molecular species of OxPLs. LC-MS/MS analysis with multiple reaction monitoring (MRM) mode has recently been used to analyze PLs and OxPLs because this method can determine acyl chain composition by monitoring a particular pair of precursor and fragment ions corresponding to the acyl chains at a specified retention time (7–13). Recently, O’Donnell and colleagues developed an excellent global lipidomic workflow that can monitor low abundant and unknown molecular species, including OxPLs (14, 15). In this procedure, untargeted lipidomics was first performed by high-resolution MS, and all detected ions were putatively identified using an online database based on m/z values without fragmentation. Then, the structural identification of selected ions was performed based on MS/MS spectra and MRM mode was applied for validation. They showed the presence of more than 100 molecular species of OxPLs by applying this procedure to the activated human platelets. However, the structure of oxidized fatty acyl chains in many molecular species of the OxPLs were not determined because of the low abundance of OxPLs generated by human platelets, and the diagnostic fragments of oxidized fatty acyl chains were hardly acquired. In this study, we first aimed to develop a precise MS/MS spectra library for OxPLs using a series of biogenic materials. Untargeted lipidomics was applied to collect MS/MS spectra for biogenic OxPLs prepared by the addition of oxidized fatty acids to HEK293 cells, where they were incorporated into cellular PLs. This procedure made it easier to identify the precise OxPL structures based on MS/MS spectra, because oxidized fatty acid was selectively incorporated into cellular PLs that produce selective OxPL molecular species. By using these MS/MS spectra for biogenic OxPLs, we successfully optimized MRM conditions and developed a broad-targeted lipidomics system to monitor about 400 molecular species of OxPLs simultaneously. This system will be useful to determine the physiological relevance of OxPLs in health and diseases.
MATERIALS AND METHODS
Materials
PLs, 1-stearoyl-2-arachidonoyl-3-PLs [phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), and phosphatidylglycerol (PG)], 1-heptadecanoyl-2-(9Z-tetradecenoyl)-sn-glycero-3-PLs [PC, PE, PI, PS, and PG], 1-stearoyl-2-linoleoyl-sn-glycero-3-PC, and 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-PE were obtained from Avanti Polar Lipids (Alabaster, AL). Oxidized fatty acids, HETE, hydroxyeicosapentaenoic acid (HEPE), hydroxydocosahexaenoic acid (HDoHE), HODE, epoxyeicosatrienoic acid (EET), epoxyeicosatetraenoic acid (EpETE), and epoxydocosapentaenoic acid (EpDPE), were obtained from Cayman Chemical (Ann Arbor, MI). Soybean 15-lipoxygenase (LOX) (type I-B), sodium borohydride, acetic acid, deoxycholate, and all solvents of LC-MS grade were obtained from Sigma-Aldrich (St. Louis, MO).
Preparation of biogenic OxPLs
HEK293 cells were cultured in DMEM supplemented with 10% FCS (Gibco, Carlsbad, CA) and 100 U/ml penicillin (Gibco), 100 μg/ml streptomycin (Gibco), and 2 mM L-glutamine (Gibco) in a 37°C incubator with 5% CO2 in air. For preparation of OxPLs, HEK293 cells (1.0 × 106 cells) were plated on a 6 cm dish and incubated with 10 μM of oxidized fatty acids for 1 h. Cells were harvested in ice-cold methanol and were extracted by solid-phase extraction using a monospin C18 column (GL Sciences, Tokyo, Japan). Briefly, the samples were applied to a monospin C18 column preconditioned with 300 μl methanol and 300 μl water and then washed with 300 μl water and 300 μl hexane followed by the elution with 250 μl isopropanol. The phosphorus content of the extracted lipids was quantified by the method of Bartlett (16); the extracted lipids were reconstituted to 1 mM phosphorus with chloroform:methanol (1:2) and then stored at −80°C until use.
Measurement of OxPL levels in primary mouse peritoneal macrophages
Male C57BL/6J mice (11 weeks) were obtained from CLEA Japan, Inc. (Tokyo, Japan). The 12/15-LOX knockout mice on a C57BL/6 background were from the Jackson Laboratory (Bar Harbor, ME). Peritoneal cells were obtained by lavage and macrophages were isolated by adherence to plastic culture plates in RPMI1640 medium supplemented with 10% FCS for 2 h at 37°C. Adherent cells were harvested and lipids were extracted by solid-phase extraction, as described above. As internal standards, PC (17:0/14:1), PE (17:0/14:1), PI (17:0/14:1), PS (17:0/14:1), and PG (17:0/14:1) were added at a final concentration of 100 nM each. The extracted lipids were reconstituted in 40 μl of chloroform:methanol (1:2) and were stored at −80°C until use. All experiments were approved by the RIKEN Yokohama Campus Animal Use Committee and performed in accordance with the committee’s guidelines.
Untargeted lipidomics
Untargeted lipidomics was performed using an ACQUITY UPLC system (Waters, Milford, MA) coupled with a quadrupole/TOF MS (TripleTOF 5600+ and TripleTOF 6600; Sciex, Framingham, MA). LC separation was performed using a reverse-phase column [ACQUITY UPLC BEH peptide C18 (50 × 1.7 mm inner diameter, 2.1 μm particle size; Waters)] with a gradient elution of mobile phase A [methanol/acetonitrile/water (1:1:3, v/v/v) containing 5 mM ammonium acetate (Wako Chemicals, Osaka, Japan) and 10 nM EDTA (Dojindo, Kumamoto, Japan)] and mobile phase B (100% isopropanol containing 5 mM ammonium acetate and 10 nM EDTA); the composition was produced by mixing those solvents. The LC gradient consisted of a holding solvent (A/B:100/0) for 1 min, then linearly converting to solvent (A/B:60/40) for 4 min, linearly converting to solvent (A/B:36/64) for 2.5 min, and holding for 4.5 min, then linearly converting to solvent (A/B:17.5/82.5) for 0.5 min, linearly converting to solvent (A/B:5/95) for 1 min followed by returning to solvent (A/B:100/0) and holding for 5 min for re-equilibration. The injection volume was 2 μl, the flow rate was 0.300 ml/min, and column temperature was 45°C. Information-dependent acquisition (IDA) mode was applied to confirm each of the biogenic OxPL structures. The source conditions were as follows: temperature, 300°C; curtain gas, 25 psi; ion source gas 1 and 2 at 80 and 40 psi; and ion spray voltage floating at −5.5 kV. The acquisition conditions were as follows: the accumulation time for full scan was 100 ms for scanning a mass range from m/z 75 to m/z 1,250. The accumulation time for each IDA experiment was 50 ms, and collision energies (CEs) were set to 35∼60 eV with a CE spread of 15 eV in high-resolution mode. IDA criteria were as follows: 10 most intense ions with an intensity threshold above 100 cps, isotope exclusion was set to 1.5 Dam, and an exclusion time of 10 s was set.
Broad-targeted analysis
Broad-targeted analysis was performed using an ACQUITY UPLC system coupled with a triple quadrupole (tripleQ) MS (QTRAP 6500; Sciex). LC separation was performed using a reverse-phase column [ACQUITY UPLC HSS T3 (50 × 2.1 mm inner diameter, 1.8 μm particle size; Waters)] with a gradient elution of mobile phase A [methanol/acetonitrile/water (1:1:3, v/v/v) containing 50 mM ammonium acetate and 10 nM EDTA] and mobile phase B (100% isopropanol containing 50 mM ammonium acetate and 10 nM EDTA); the composition was produced by mixing those solvents. The LC gradient consisted of holding solvent (A/B:100/0) for 1 min, then linearly converting to solvent (A/B:50/50) for 4 min, linearly converting to solvent (A/B:36/64) for 7 min, then linearly converting to solvent (A/B:5/95) for 1 min and holding for 1 min followed by returning to solvent (A/B:100/0) and holding for 5 min for re-equilibration. The injection volume was 3.5 μl, the flow rate was 0.350 ml/min, and column temperature was 50°C. MRM mode was applied to detection of OxPLs in biological samples. Selected MRM transitions and CEs are described in Table 1 and supplemental Table S1. For quantification, OxPL standard solutions corresponding to 10, 20, 50, 100, 200, and 500 nM were prepared to acquire calibration curves for concentration and efficiency of ionization. One microliter of those solutions was injected and measured as described above. Calibration curves were obtained from the concentrations and the area of intensity of each OxPL.
TABLE 1.
Representative optimized MRM transitions for OxPLs detected by untargeted lipidomics
| Head | Adduct Ion | sn-1 | sn-2 | Q1 (m/z) | Q3 (m/z) | CE (eV) | Retention Time (min) |
| PC | [M+CH3COO]− | 16:0 | 5-HETE | 856.6 | 115.0 | −47 | 9.2 |
| 8-HETE | 856.6 | 155.1 | −47 | 8.7 | |||
| 12-HETE | 856.6 | 179.1 | −50 | 8.5 | |||
| 15-HETE | 856.6 | 219.1 | −50 | 8.2 | |||
| 18-HETE | 856.6 | 261.2 | −56 | 7.8 | |||
| 20-HETE | 856.6 | 289.2 | −56 | 7.7 | |||
| 5-HEPE | 854.6 | 115.0 | −46 | 8.6 | |||
| 8-HEPE | 854.6 | 155.1 | −52 | 8.1 | |||
| 15-HEPE | 854.6 | 219.1 | −49 | 7.5 | |||
| 7-HDoHE | 880.6 | 141.1 | −47 | 8.8 | |||
| 10-HDoHE | 880.6 | 153.1 | −53 | 8.4 | |||
| 17-HDoHE | 880.6 | 245.2 | −47 | 8.2 | |||
| 20-HDoHE | 880.6 | 285.2 | −44 | 8.0 | |||
| 14,15-EET | 856.6 | 219.1 | −47 | 8.8 | |||
| 8,9-EpETE | 854.6 | 155.1 | −41 | 8.6 | |||
| 11,12-EpETE | 854.6 | 179.1 | −47 | 8.4 | |||
| 14,15-EpETE | 854.6 | 219.1 | −47 | 8.3 | |||
| 17,18-EpETE | 854.6 | 259.2 | −44 | 8.1 | |||
| 19,20-EpDPE | 880.6 | 285.2 | −44 | 8.6 | |||
| 13-HODE | 832.6 | 195.1 | −59 | 7.9 |
Preparation of purified OxPL standards by soybean 15-LOX
OxPL standards were generated in accordance with a previous method (10). Briefly, PC (18:0/20:4), PE (18:0/20:4), PI (18:0/20:4), PS (18:0/20:4), and PG (18:0/20:4) were incubated with soybean 15-LOX in 0.2 M borate buffer (pH 9.0) and 10 mM deoxycholate at room temperature. The oxidation reaction was monitored by its absorbance at 234 nm and was terminated at 30 min. Hydroperoxide intermediates were reduced with excess sodium borohydride and incubations were extracted by using a monospin C18 column as described above. To separate and isolate conversion products, reverse-phase HPLC was carried out by using a Waters XBridge C18 column (100 × 4.6 mm inner diameter, 5 μm particle size; Waters) with mobile phase A [methanol/water (1:1) containing 0.01% acetic acid] and mobile phase B (100% methanol) at 0.7 ml/min of flow rate for PC and PE, and an ACQUITY UPLC HSS T3 (75 × 3.5 mm inner diameter, 1.8 μm particle size; Waters) with mobile phase A [methanol/acetonitrile/water (1:1:3, v/v/v) containing 50 mM ammonium acetate and 10 nM EDTA] and mobile phase B (100% isopropanol containing 50 mM ammonium acetate and 10 nM EDTA) at 0.6 ml/min for PI, PG, and PS. The concentration of purified standards was assessed by LC-MS/MS after saponification with 0.2 M NaOH at 60°C for 30 min in 50% isopropanol.
Preparation of positional isomer mixtures of OxPL standards by air oxidation
Positional isomer mixtures of OxPL standards were generated by air oxidation in accordance with a previous method (10). Briefly, 10 mM 2,2,5,7,8-pentamethyl-6-chromanol (Sigma-Aldrich) were added to PC (18:0/20:4), PE (18:0/20:4), PE (18:0/22:6), and PC (18:0/18:2), and they were dried up under nitrogen gas. Then, the samples were incubated at 37°C for 24 h followed by resuspension in methanol containing 20 mg/ml butylated hydroxytoluene (Sigma-Aldrich). Lipids were extracted by using a monospin C18 column, as described above. To purify conversion products, reverse-phase HPLC was carried out by using a Waters XBridge C18 column (100 × 4.6 mm inner diameter, 5 μm particle size; Waters) with mobile phase A [methanol/acetonitrile/water (1:1:3, v/v/v) containing 0.1% acetate and 10 nM EDTA] and mobile phase B (100% isopropanol containing 0.1% acetate and 10 nM EDTA) at 1.0 ml/min. The concentration of purified standards was assessed by LC-MS/MS after saponification with 0.2 M NaOH at 60°C for 30 min in 50% isopropanol.
Statistical analysis
Results are expressed as mean ± SE. Differences between two groups were tested by the Student’s t-test. A significance level of P < 0.05 was used.
RESULTS
Construction of a measured MS/MS spectra library for OxPLs
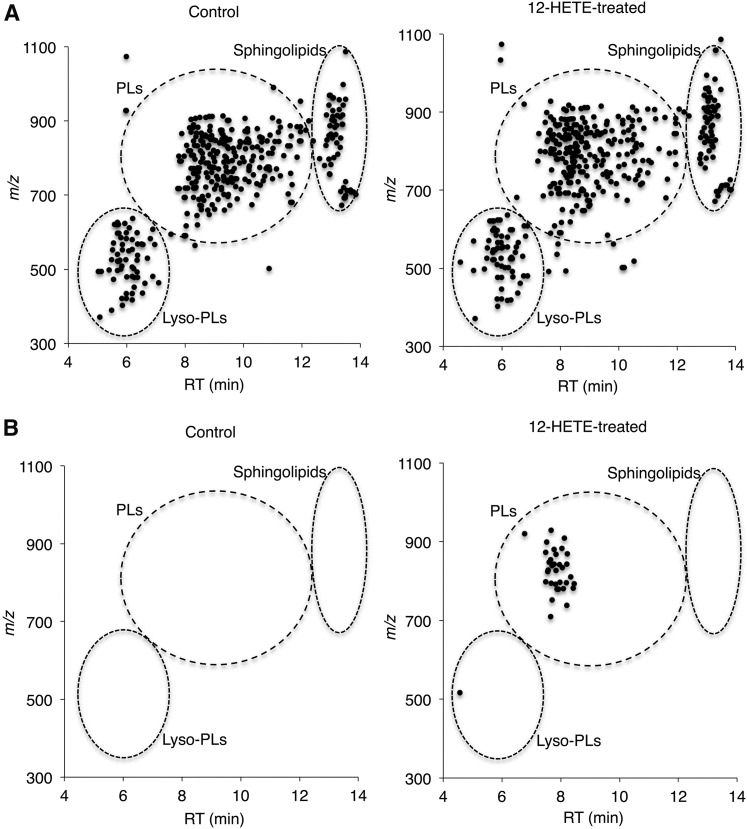
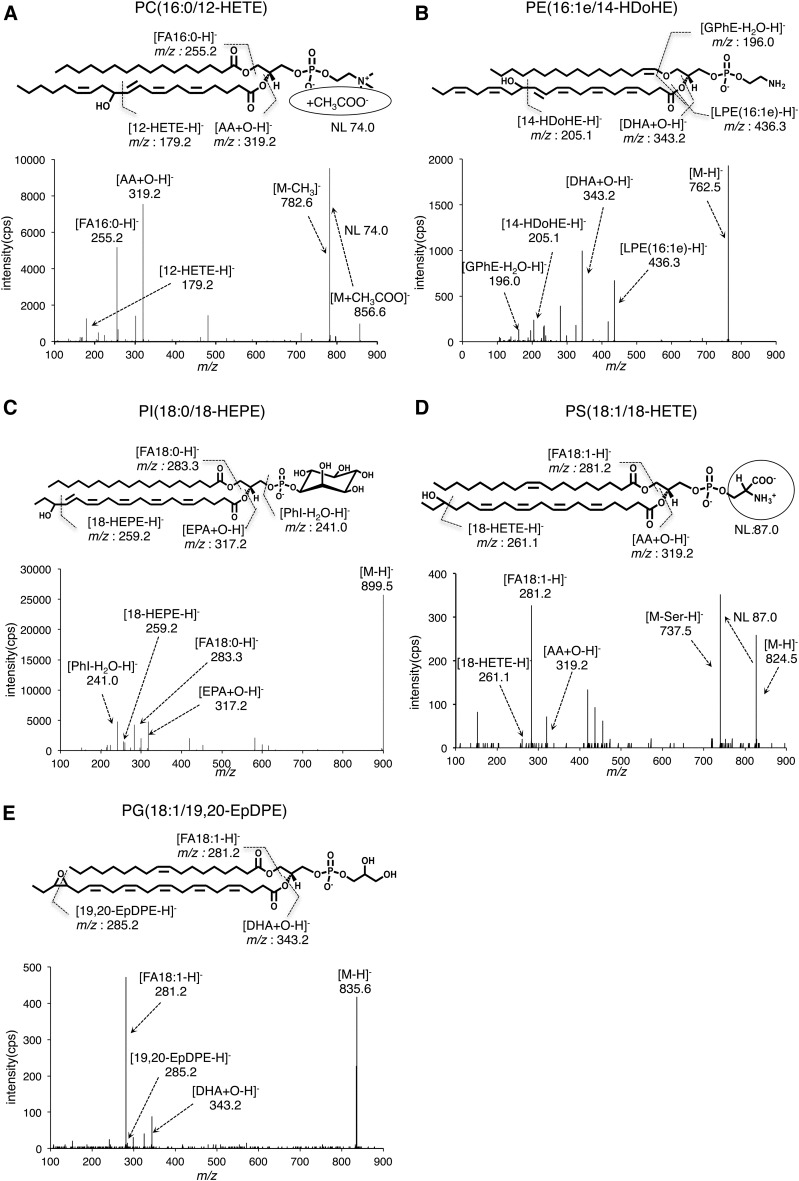
To acquire MS/MS spectra for OxPLs, we devised a method to prepare various types of OxPLs by use of biogenic conversion from oxidized fatty acids incorporated into cellular PLs. Oxidized fatty acids, such as hydroxyl and epoxy-containing fatty acids, were added to HEK293 cells for 1 h, cells were harvested, and lipids were extracted. Lipid fractions were analyzed by LC-quadrupole/TOF (QTOF)-MS-based untargeted lipidomics to collect MS/MS spectra for each of the biogenic OxPLs. This method provides automatic MS to MS/MS switching by setting the MS/MS trigger at a low threshold level of intensity and then information-rich MS/MS spectra with high resolution could be acquired in a nonbiased fashion (17–19). For example, many types of lipids, such as lyso-PLs, PLs, and sphingolipids, were readily detected in lipid extracts of HEK293 cells and the candidate signals for PLs containing 12-HETE were obtained in 12-HETE-treated cells, as determined by the presence of a fragment ion (m/z 319.2) corresponding to mono-oxidized AA (abbreviated as AA+O) (Fig. 1). MS/MS data of those candidate signals were further carefully investigated and diagnostic fragment ions for accurate identification were determined. For example, PC (16:0/12-HETE) was identified by the precursor ion (m/z 856.5) and fragment ions of acyl chains (m/z 255.2→16:0, m/z 319.2→AA+O), neutral loss ion of the choline part ([M+CH3COO-74]−), and 12-HETE-specific fragment ion (m/z 179.1), which could distinguish it from other HETEs (Fig. 2). In total, 12 types of OxPC, 8 types of OxPE, 3 types of OxPI, and 2 types of OxPS were identified in extracts from 12-HETE-treated cells (Table 1). This procedure was also applied to other oxidized fatty acids, i.e., HETEs, HEPEs, HDoHEs, HODEs, EETs, EpETEs, and EpDPEs, in order to expand the MS/MS spectra library. Characteristic fragment ions for identification of those OxPLs were individually optimized and MS/MS data of 386 total molecular species were successfully acquired (Fig. 2, Table 1, supplemental Table S1). Full MS/MS spectra data are deposited in the “Computational MS-based metabolomics” section of the RIKEN PRIMe website (http://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL/index.html).
Fig. 1.
2D map of mass versus LC retention time of lipids isolated from control and 12-HETE-treated cells. The 2D map has the m/z values of precursor ions along the vertical axis and the retention times (RT) along the horizontal axis. A: Plots of precursor ions identified as lyso-PLs, PLs, and sphingolipids. B: Plots of candidate signals for PLs containing 12-HETE determined by the presence of a fragment ion (m/z 319.2) corresponding to AA+O.
Fig. 2.
Structural identification of OxPLs by MS/MS fragmentation patterns. Structural analysis of candidate signals obtained from a series of biogenic OxPLs. Based on the MS/MS spectra, the OxPL structures were assigned to the corresponding fragments of the acyl chains, the diagnostic ions of molecular species of oxidized fatty acids, and the head groups or neutral loss ions of the head groups. GPhE, phosphoglycerol ethanolamine; LPE, lyso-PE; PhI, phosphoinositol; NL, neutral loss.
Development of a specific and highly sensitive analytical system for OxPLs
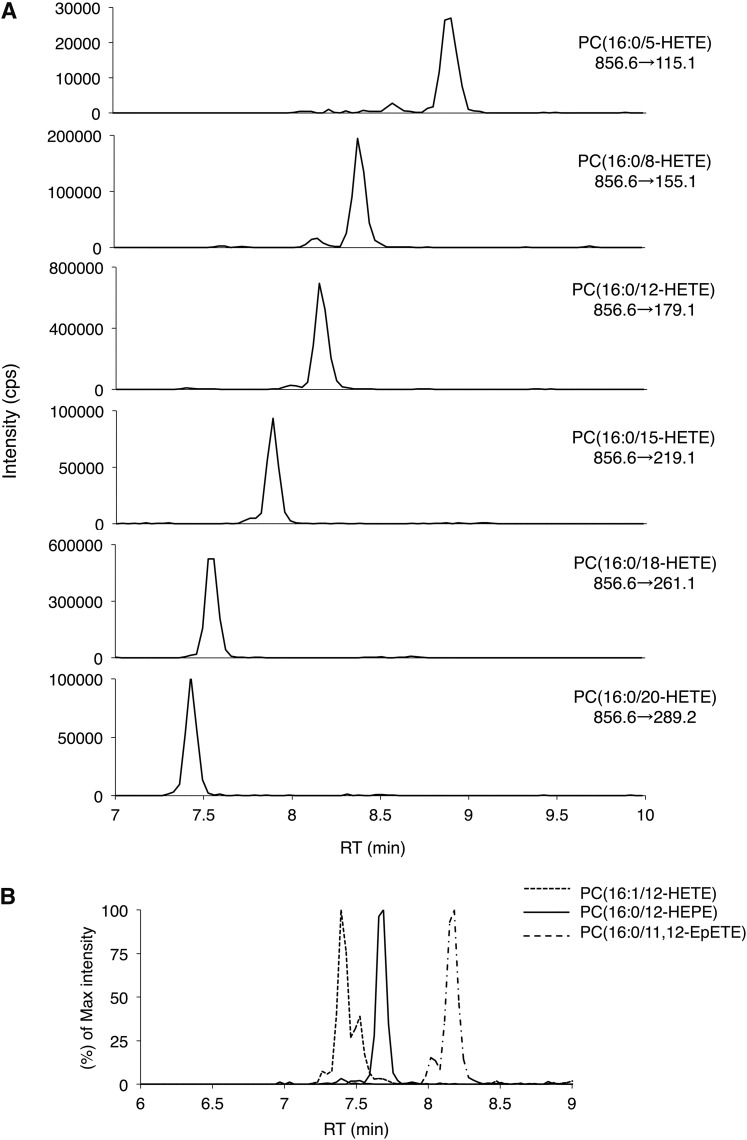
The abundance of OxPLs in biological samples is normally low and many of them are short-lived and structurally similar (9–15). Therefore, the development of a highly specific and sensitive method to monitor OxPL molecular species in vivo is important to understand their physiological and/or pathophysiological roles. MRM analysis with a particular pair of precursor and fragment ions at a specified retention time using LC tripleQ MS was applied to demonstrate broad-targeted analysis of OxPLs. Based on the fragmentation patterns of MS/MS spectra library, MRM transitions from a targeted precursor ion to its specific fragment ion were carefully selected to avoid peak overlapping for individual OxPL molecular species. For example, structural differences of OxPLs, such as PC (16:0/5-HETE), PC (16:0/8-HETE), PC (16:0/12-HETE), and PC (16:0/15-HETE), were distinguished by the optimized MRM transitions, as depicted in Fig. 3A. In the case of OxPLs whose MS/MS spectra could not be acquired by untargeted lipidomics, we predicted fragmentation patterns for them to expand the measurement range as predictive MRM. LC gradient conditions were also optimized to separate similar OxPL structural isomers (Fig. 3B). The CE was optimized by determining the most sensitive value for each biogenic OxPL individually. The optimized MRM conditions for each OxPL molecular species are summarized in Table 1 and supplemental Table S1.
Fig. 3.
OxPL detection by LC tripleQ MS with optimized MRM transitions and LC gradient conditions. A: A series of biogenic OxPCs containing HETEs were selectively detected by optimized MRM transitions. B: Structural isomers were separated by the optimized LC gradient condition.
Preparation of purified OxPL standards for determination of the linear ranges and the lower limits of detection
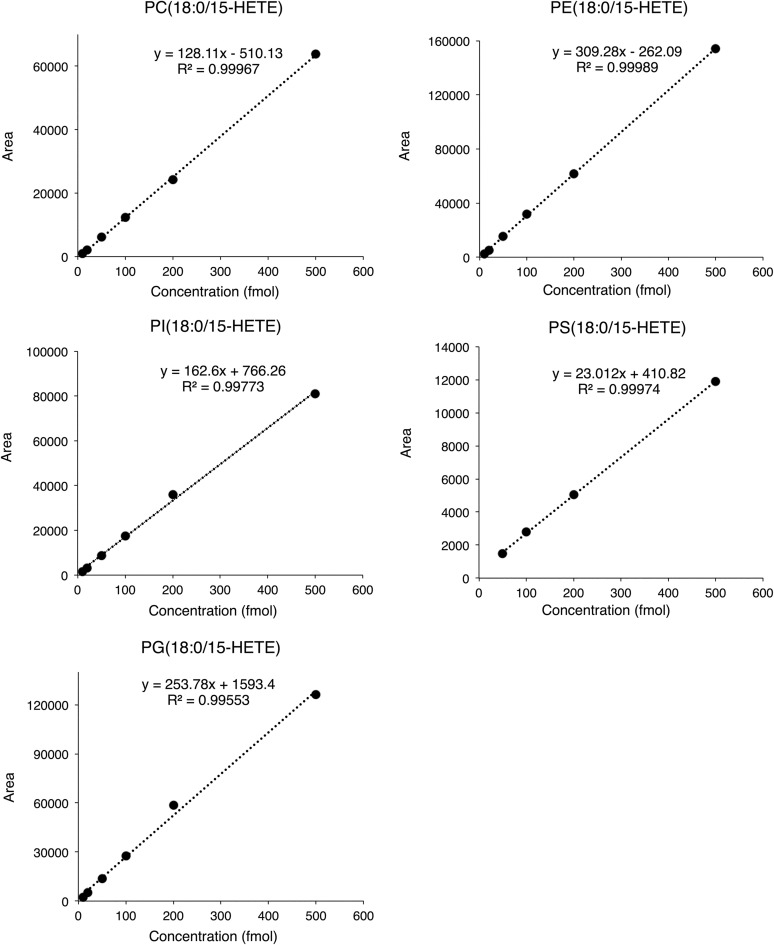
To determine the linear ranges and the lower limits of detection in our analytical system, purified standard compounds were prepared using an in vitro LOX reaction. OxPLs containing 15-HETE as representative OxPL standards were prepared by in vitro oxidation of PC (18:0/20:4), PE (18:0/20:4), PI (18:0/20:4), PS (18:0/20:4), and PG (18:0/20:4) using soybean 15-LOX. The concentrations of the OxPL standards were assessed by measuring the amount of oxidized fatty acids after saponification (20, 21). The peak areas were acquired by OxPC, OxPE, OxPI, OxPS, and OxPG standard solutions corresponding to 10, 20, 50, 100, 200, and 500 nM (Fig. 4). The linear ranges were between 10 and 500 fmol for OxPC, OxPE, OxPI, and OxPG and between 50 and 500 fmol for OxPS. These results suggested that the lower limits of detection in our OxPL lipidomics system were 10 fmol for OxPC, OxPE, OxPI, and OxPG and 50 fmol for OxPS on column (19–21).
Fig. 4.
Calibration curves for quantification of OxPLs. OxPL standard solutions corresponding to 10, 20, 50, 100, 200, and 500 fmol were quantified by LC tripleQ MS with optimized MRM transitions. Calibration curves were constructed by plotting the peak areas and concentrations.
Application of the OxPL lipidomics system to a biological sample
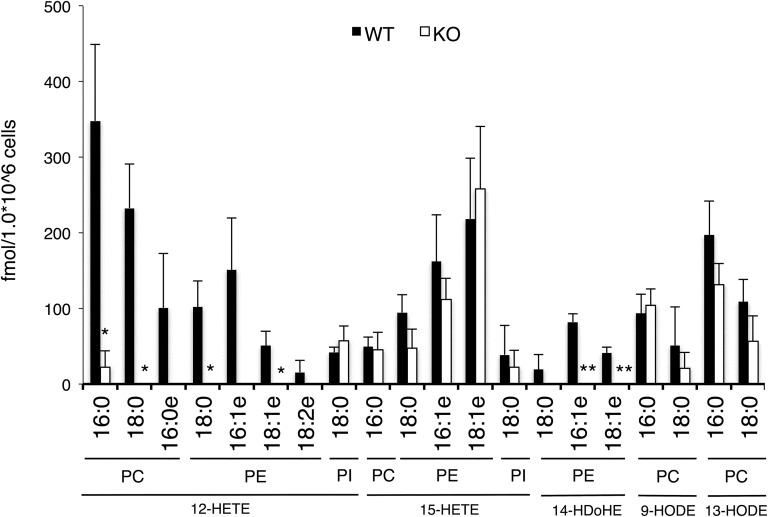
To determine whether this OxPL lipidomics system was applicable to biological samples, we applied it to mouse peritoneal macrophages. Previously, it was reported that mouse peritoneal macrophages express a cytosolic LOX, 12/15-LOX, and contain a series of OxPLs endogenously produced by the 12/15-LOX activity (11). Lipids were extracted from mouse peritoneal macrophages and the lipid fractions were applied to LC-MS/MS-based OxPL lipidomics with optimized MRM conditions. The extraction efficiency was normalized using PC (17:0/14:1), PE (17:0/14:1), and PI (17:0/14:1) as internal standards. Using this approach, 20 molecular species of OxPLs containing 12-HETE, 15-HETE, 14-HDoHE, 9-HODE, and 13-HODE were successfully detected (Fig. 5). To quantify the amount of those OxPLs, we prepared positional isomer mixtures of OxPL standards by air oxidation of PC (18:0/20:4), PE (18:0/20:4), PE (18:0/22:6), and PC (18:0/18:2), and constructed calibration curves (supplemental Fig. S1). As a result of quantification using these calibration curves, the total amounts of OxPC, OxPE, and OxPI were estimated to be 1180.3, 936.5, and 80.7 fmol per 1.0 × 106 macrophages, respectively. These results were consistent with the previous report (11). Also, a series of OxPLs containing 12/15-LOX products, such as 12-HETE and 14-HDoHE, were clearly detected in WT macrophages, but were much less abundant in 12/15-LOX-deficient macrophages (Fig. 5). These results suggest that our LC-MS/MS-based lipidomics system is sensitive enough to monitor individual OxPL molecular species present in biological samples.
Fig. 5.
Quantification of OxPLs in mouse peritoneal macrophages. Peritoneal macrophages isolated from WT and 12/15-LOX KO mice were analyzed. Levels of OxPLs were expressed as concentration per 1.0 × 106 cells. (n = 3, mean ± SE, *P < 0.05, **P < 0.01).
DISCUSSION
In this study, we first constructed a measured MS/MS spectra library by applying untargeted lipidomics to biogenic OxPLs prepared by the addition of oxidized fatty acids to cultured cells. Based on the MS/MS spectra for each OxPL molecular species, we developed a broad-targeted lipidomics system by MRM mode. We applied this system to mouse peritoneal macrophages and quantified a series of OxPLs endogenously produced in a 12/15-LOX-dependent manner. These results indicate that our lipidomics system will be applicable to the comprehensive analysis of OxPLs present in biological samples.
The previous analytical limitations were due to the lack of various types of OxPL standards. To solve this problem, we conceived the idea to prepare various types of OxPLs by adding different types of oxidized fatty acids to HEK293 cells, because these exogenous oxidized fatty acids would be incorporated into the cellular PLs by lyso-PL acyltransferase (22, 23). In this study, a total of 29 types of oxidized fatty acids were added to HEK293 cells and about 400 molecular species of OxPLs were identified by untargeted lipidomics. Interestingly, some of these OxPLs had unique incorporation patterns for PLs. For example, 15-HETE and 18-HEPE were effectively incorporated into PI; whereas, 12-HETE was preferentially incorporated into PC. Those preferences for oxidized fatty acids may depend on the substrate specificities of the lyso-PL acyltransferases. Therefore, it might be possible to expand the biogenic MS/MS spectra library by manipulating these enzyme activities.
We also found that the LC retention time for OxPLs appeared to be affected by the hydroxyl and epoxy group position in the oxidized fatty acyl chains. For example, epoxy-containing OxPLs eluted later than hydroxyl-containing OxPLs, and OxPLs whose hydroxyl or epoxy group was located closer to the end of acyl chain eluted earlier than other OxPLs. Also, the optimal CE condition appeared to be dependent on the PL head group rather than on the molecular species of oxidized fatty acyl chains (Table 1, supplemental Table S1).
A previous study reported that 12/15-LOX-dependent OxPL production by mouse peritoneal macrophages maintains immunogenic tolerance by interfering with MFG-E8-mediated uptake of apoptotic cells into inflammatory monocytes (24). A series of OxPLs containing 12-HETE, 15-HETE, and 14-HDoHE were identified in those macrophages. OxPLs are also formed in other cell types, such as neutrophils, platelets, and dendritic cells (DCs) (25–28). These OxPLs are thought to have various physiological functions, such as production of neutrophil extracellular traps, thrombin generation of platelets, and regulation of DC maturation and function (25, 26, 28). Application of the present method to those biological samples would enable us to monitor the structure-specific behavior of OxPLs that could lead to the elucidation of their physiological relevance.
In summation, we constructed a broad-targeted lipidomics system for OxPLs based on the fragmentation patterns of a measured MS/MS spectra library. This lipidomics system will enable us to monitor OxPLs comprehensively and will help us to understand their physiological significance in a quantitative and qualitative manner.
Supplementary Material
Acknowledgments
The authors thank Y. Senoo for technical assistance.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- AA+O
- mono-oxidized arachidonic acid
- CE
- collision energy
- DC
- dendritic cell
- EET
- epoxyeicosatrienoic acid
- EpDPE
- epoxydocosapentaenoic acid
- EpETE
- epoxyeicosatetraenoic acid
- HDoHE
- hydroxydocosahexaenoic acid
- HEPE
- hydroxyeicosapentaenoic acid
- IDA
- information-dependent acquisition
- LC-QTOF-MS
- LC-quadrupole/TOF-MS
- LOX
- lipoxygenase
- MRM
- multiple reaction monitoring
- OxPL
- oxidized phospholipid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PL
- phospholipid
- PS
- phosphatidylserine
- tripleQ
- triple quadrupole
This work was supported by Japan Society for the Promotion of Science Grants 15H05898, 15H05897, and 15H04648; the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry; the Japan Agency for Medical Research and Development, and the Japan Science and Technology Agency Core Research for Evolutional Science and Technology. Additional support was provided by the RIKEN Junior Research Associate Program (to R.A.).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Reis A., and Spickett C. M.. 2012. Chemistry of phospholipid oxidation. Biochim. Biophys. Acta. 1818: 2374–2387. [DOI] [PubMed] [Google Scholar]
- 2.Ashraf M. Z., Kar N. S., and Podrez E. A.. 2009. Oxidized phospholipids: biomarker for cardiovascular diseases. Int. J. Biochem. Cell Biol. 41: 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsuzura S., Ikeda Y., Suehiro T., Ota K., Osaki F., Arii K., Kumon Y., and Hashimoto K.. 2004. Correlation of plasma oxidized low-density lipoprotein levels to vascular complications and human serum paraoxonase in patients with type 2 diabetes. Metabolism. 53: 297–302. [DOI] [PubMed] [Google Scholar]
- 4.Hammad L. A., Wu G., Saleh M. M., Klouckova I., Dobrolecki L. E., Hickey R. J., Schnaper L., Novotny M. V., and Mechref Y.. 2009. Elevated levels of hydroxylated phosphocholine lipids in the blood serum of breast cancer patients. Rapid Commun. Mass Spectrom. 23: 863–876. [DOI] [PubMed] [Google Scholar]
- 5.Huang L. S., Kim M. R., Jeong T. S., and Sok D. E.. 2007. Linoleoyl lysophosphatidic acid and linoleoyl lysophosphatidylcholine are efficient substrates for mammalian lipoxygenases. Biochim. Biophys. Acta. 1770: 1062–1070. [DOI] [PubMed] [Google Scholar]
- 6.Thomas C. E., Ku G., and Kalyanaraman B.. 1994. Nitrone spin trap lipophilicity as a determinant for inhibition of low density lipoprotein oxidation and activation of interleukin-1 beta release from human monocytes. J. Lipid Res. 35: 610–619. [PubMed] [Google Scholar]
- 7.Nakanishi H., Ogiso H., and Taguchi R.. 2009. Qualitative and quantitative analyses of phospholipids by LC-MS for lipidomics. Methods Mol. Biol. 579: 287–313. [DOI] [PubMed] [Google Scholar]
- 8.Myers D. S., Ivanova P. T., Milne S. B., and Brown H. A.. 2011. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim. Biophys. Acta. 1811: 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakanishi H., Iida Y., Shimizu T., and Taguchi R.. 2009. Analysis of oxidized phosphatidylcholines as markers for oxidative stress, using multiple reaction monitoring with theoretically expanded data sets with reversed-phase liquid chromatography/tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 1366–1374. [DOI] [PubMed] [Google Scholar]
- 10.Morgan A. H., Hammond V. J., Morgan L., Thomas C. P., Tallman K. A., Garcia-Diaz Y. R., McGuigan C., Serpi M., Porter N. A., Murphy R. C., et al. . 2010. Quantitative assays for esterified oxylipins generated by immune cells. Nat. Protoc. 5: 1919–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan A. H., Dioszeghy V., Maskrey B. H., Thomas C. P., Clark S. R., Mathie S. A., Lloyd C. M., Kühn H., Topley N., Coles B. C., et al. . 2009. Phosphatidylethanolamine-esterified eicosanoids in the mouse: tissue localization and inflammation-dependent formation in Th-2 disease. J. Biol. Chem. 284: 21185–21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchikata T., Matsubara A., Nishiumi S., Yoshida M., Fukusaki E., and Bamba T.. 2012. Development of oxidized phosphatidylcholine isomer profiling method using supercritical fluid chromatography/tandem mass spectrometry. J. Chromatogr. A. 1250: 205–211. [DOI] [PubMed] [Google Scholar]
- 13.Reis A., Domingues P., and Domingues M. R.. 2013. Structural motifs in primary oxidation products of palmitoyl-arachidonoyl-phosphatidylcholines by LC-MS/MS. J. Mass Spectrom. 48: 1207–1216. [DOI] [PubMed] [Google Scholar]
- 14.Slatter D. A., Aldrovandi M., O’Connor A., Allen S. M., Brasher C. J., Murphy R. C., Mecklemann S., Ravi S., Darley-Usmar V., and O’Donnell V. B.. 2016. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metab. 23: 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connor A., Brasher C. J., Slatter D. A., Meckelmann S. W., Hawksworth J. I., Allen S. M., and O’Donnell V. B.. 2017. LipidFinder: a computational workflow for discovery of lipids identifies eicosanoid-phosphoinositides in platelets. JCI Insight. 2: e91634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlett G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234: 466–468. [PubMed] [Google Scholar]
- 17.Decaestecker T. N., Clauwaert K. M., Van Bocxlaer J. F., Lambert W. E., Van den Eeckhout E. G., Van Peteghem C. H., and De Leenheer A. P.. 2000. Evaluation of automated single mass spectrometry to tandem mass spectrometry function switching for comprehensive drug profiling analysis using a quadrupole time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 14: 1787–1792. [DOI] [PubMed] [Google Scholar]
- 18.Decaestecker T. N., Vande Casteele S. R., Wallemacq P. E., Van Peteghem C. H., Defore D. L., and Van Bocxlaer J. F.. 2004. Information-dependent acquisition-mediated LC-MS/MS screening procedure with semiquantitative potential. Anal. Chem. 76: 6365–6373. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda K. 2015. Mass-spectrometric analysis of phospholipids by target discovery approach. In Yokomizo T. and Murakami M., editors. Springer, Japan: 349–356. [Google Scholar]
- 20.Yamada T., Tani Y., Nakanishi H., Taguchi R., Arita M., and Arai H.. 2011. Eosinophils promote resolution of acute peritonitis by producing proresolving mediators in mice. FASEB J. 25: 561–568. [DOI] [PubMed] [Google Scholar]
- 21.Isobe Y., Arita M., Matsueda S., Iwamoto R., Fujihara T., Nakanishi H., Taguchi R., Masuda K., Sasaki K., Urabe D., et al. . 2012. Identification and structure determination of novel anti-inflammatory mediator resolvin E3, 17,18-dihydroxyeicosapentaenoic acid. J. Biol. Chem. 287: 10525–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brezinski M. E., and Serhan C. N.. 1990. Selective incorporation of (15S)-hydroxyeicosatetraenoic acid in phosphatidylinositol of human neutrophils: agonist-induced deacylation and transformation of stored hydroxyeicosanoids. Proc. Natl. Acad. Sci. USA. 87: 6248–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shindou H., and Shimizu T.. 2009. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 284: 1–5. [DOI] [PubMed] [Google Scholar]
- 24.Uderhardt S., Herrmann M., Oskolkova O. V., Aschermann S., Bicker W., Ipseiz N., Sarter K., Frey B., Rothe T., Voll R., et al. . 2012. 12/15-lipoxygenase orchestrates the clearance of apoptotic cells and maintains immunologic tolerance. Immunity. 36: 834–846. [DOI] [PubMed] [Google Scholar]
- 25.Clark S. R., Guy C. J., Scurr M. J., Taylor P. R., Kift-Morgan A. P., Hammond V. J., Thomas C. P., Coles B., Roberts G. W., Eberl M., et al. . 2011. Esterified eicosanoids are acutely generated by 5-lipoxygenase in primary human neutrophils and in human and murine infection. Blood. 117: 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas C. P., Morgan L. T., Maskrey B. H., Murphy R. C., Kühn H., Hazen S. L., Goodall A. H., Hamali H. A., Collins P. W., and O’Donnell V. B.. 2010. Phospholipid-esterified eicosanoids are generated in agonist-activated human platelets and enhance tissue factor-dependent thrombin generation. J. Biol. Chem. 285: 6891–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan L. T., Thomas C. P., Kühn H., and O’Donnell V. B.. 2010. Thrombin-activated human platelets acutely generate oxidized docosahexaenoic-acid-containing phospholipids via 12-lipoxygenase. Biochem. J. 431: 141–148. [DOI] [PubMed] [Google Scholar]
- 28.Rothe T., Gruber F., Uderhardt S., Ipseiz N., Rössner S., Oskolkova O., Blüml S., Leitinger N., Bicker W., Bochkov V. N., et al. . 2015. 12/15-Lipoxygenase-mediated enzymatic lipid oxidation regulates DC maturation and function. J. Clin. Invest. 125: 1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.