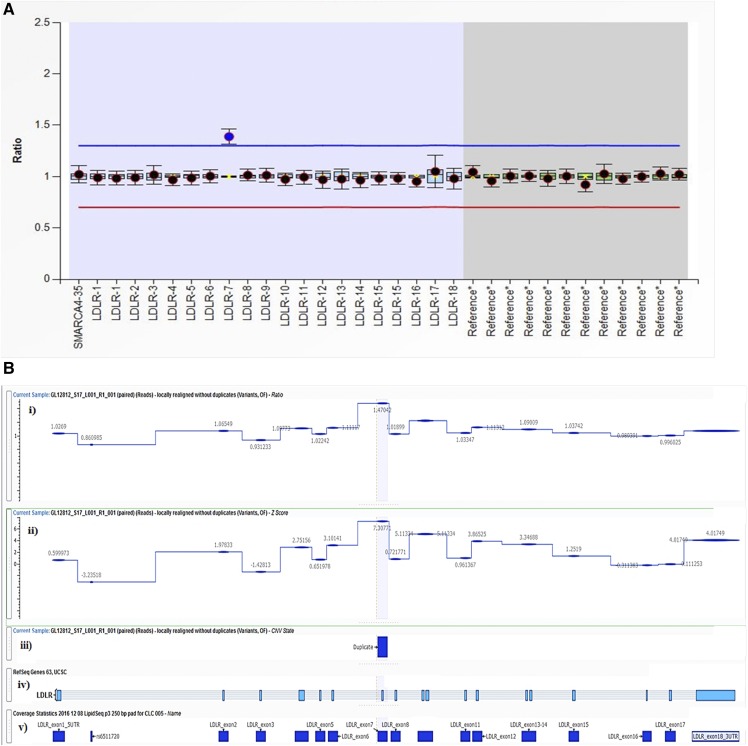
Fig. 2.
Two methods of detection of a CNV duplication event in the LDLR gene in a patient with FH (subject GL12812) with a duplication in LDLR exon 7. See Fig. 1 legend for overall structure of the panels. A: MLPA method output: duplication in LDLR exon 7. B: VarSeq CNV Caller method output: duplication in LDLR exon 7. i: Normalized ratio metric computed for each LipidSeq target region in LDLR; depth of sequence coverage comparative to reference controls where ∼1.0 indicates diploid (normal) copy number state and ∼1.5 indicates a duplication event. ii: Normalized z-score metric; number of SDs the DOC is from the reference control mean coverage, where ≥5.0 is the threshold set to indicate a duplication event; other sections are as in Fig. 1.