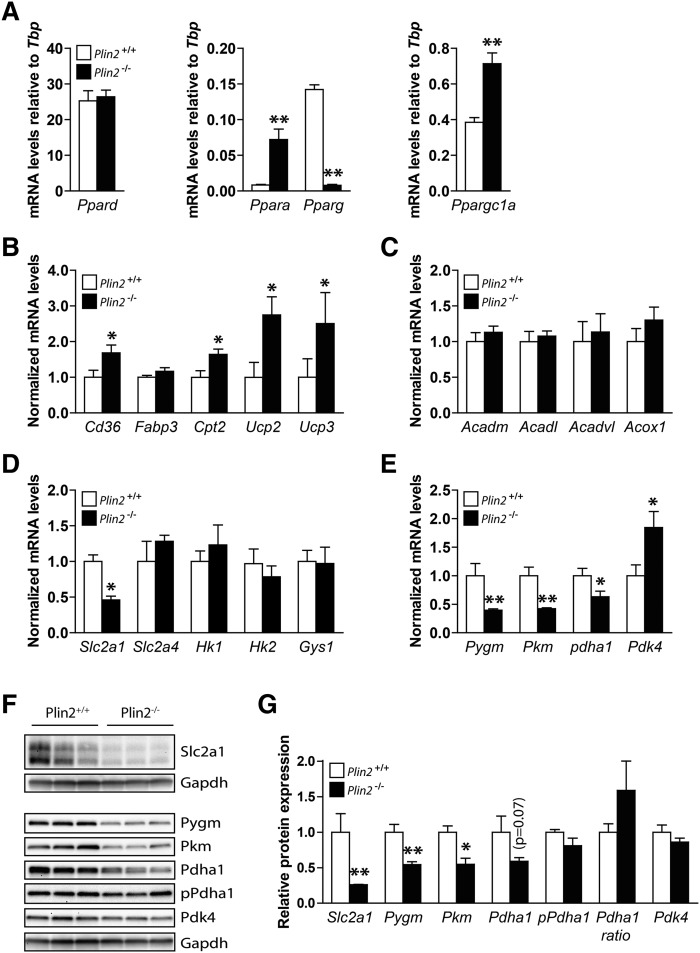
Fig. 6.
Expression of genes involved in lipid metabolism in Plin2+/+ and Plin2−/− myotubes. Gene expression of various mRNAs in differentiated Plin2+/+ and Plin2−/− myotubes was analyzed by RT-qPCR and related to the expression of TATA-box binding protein (Tbp). A: Expression of transcription factors activated by FAs; Ppar α, γ and δ and Ppar γ coactivator 1 α (Ppargc1a). Results are presented as means ± SEM (n = 4, *P < 0.05 and **P < 0.01 vs. Plin2+/+). B: Expression of genes involved in lipid uptake and mitochondrial function; FA transporter/CD36 antigen (Cd36), Fabp3, Cpt2, and uncoupling proteins 2 and 3 (Ucp2 and Ucp3, respectively). C: Expression of genes catalyzing oxidation of FAs; mitochondrial Acadm, Acadl, Acadvl, and peroxisomal Acox1. D: Expression of genes involved in glucose uptake and storage; solute carrier family 2 member 1 and 4 (Slc2a1 and Slc2a4, respectively), hexokinase 1 and 2 (Hk1 and Hk2, respectively) and Gys1. E: Expression of genes involved in glycogen mobilization and glucose oxidation; muscle-associated glycogen phosphorylase(Pygm), muscle pyruvate kinase (Pkm), pyruvate dehydrogenase α 1 (Pdha1), and pyruvate dehydrogenase kinase 4 (Pdk4). Results in B–F are presented as means ± SEM normalized to the expression in Plin2+/+ myotubes (n = 5, *P < 0.05, **P < 0.01 vs. Plin2+/+). F: Protein content of Slc2a1, Pygm, Pkm, Pdha1, Pdha1 phosphorylated at Ser300 (pPdha1), and Pdk4 (n = 3). G: Protein content, related to Gapdh normalized to the expression levels in Plin2+/+ myotubes, or the ratio of pPdha1 against total Pdha1 (Pdha1 ratio). The results are presented as means ± SEM (n = 3, *P < 0.05 vs. Plin2+/+).