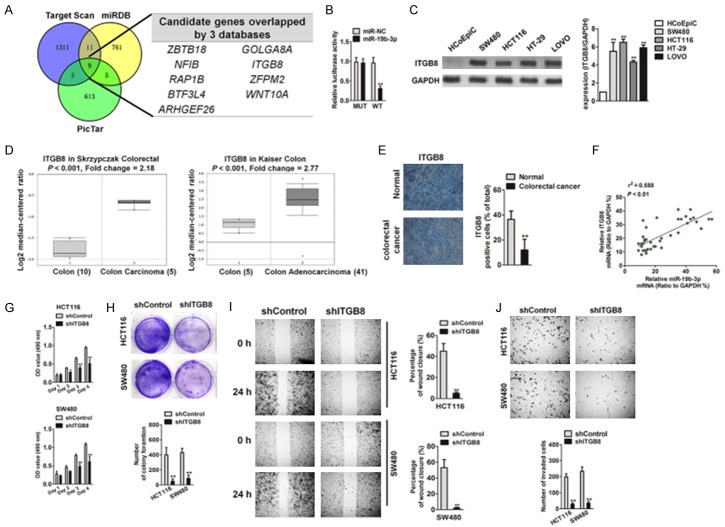
Figure 5.
Effect of ITGB8 knockdown on CRC cells growth and metastasis. A. miR19b-3p target genes were predicted by in silico analyses. The Venn graph represented the number of candidate genes determined by three prediction algorithms. B. The relative luciferase activity in 293T cells co-transfected with the miR19b-3p mimic and WT vector or MUT vector was analyzed by luciferase reporter assay. C. RNA was extracted from CRC cells and the levels of ITGB8 were determined by qRT-PCR (upper panel). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, compared with HCoEpiC cells. The expression of ITGB8 was detected by western blotting of CRC cell lines (lower panel). D. ITGB8 was overexpressed in CRC tissues compared to that in normal tissues, based on the datasets from Oncomine. E. ITGB8 levels in CRC tissues were measured by IHC. F. The positive correlation analysis of miR19b-3p and ITGB8 in CRC tissues. G. Cell proliferation activity was measured by MTT assay. H. The growth capacity was determined in the colony formation assay. I. HCT116 and SW480 cell migration was analyzed by the wound healing assay as described above. J. HCT116 and SW480 cell invasion capacity was analyzed by Transwell Matrigel invasion assay as described above. Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, compared with control cells.