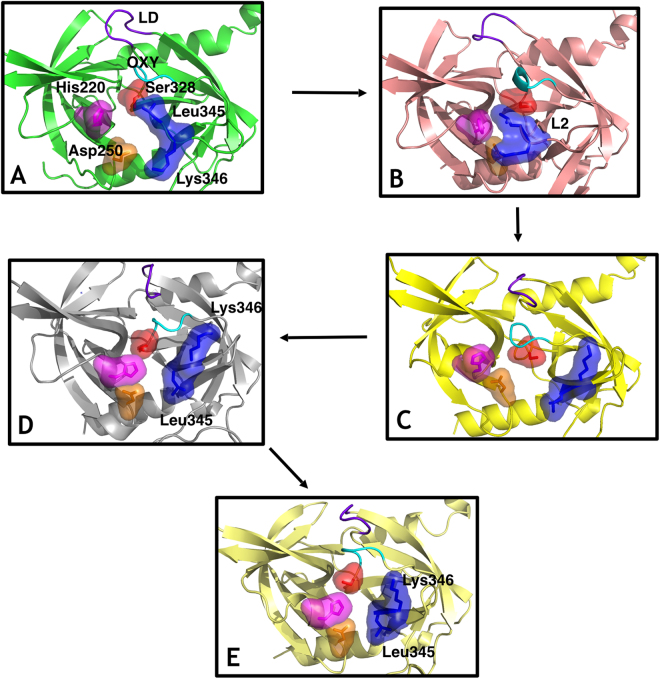
Figure 1.
Sequence of states in the activation process from computational simulation. The residues Ser328 (surface, red), His220 (surface, magenta), Asp250 (surface, orange), L2 loop residues Leu345 and Lys346 (surface, blue), the LD loop (purple) and the oxyanion hole forming loop (cyan) are highlighted. (A) Fully inactive state that closely resembles the X-ray inactive structure (PDB 3NUM). The misaligned configuration of the catalytic triad (Ser328, His220 and Asp250) is incompatible with catalytic activity. In the binding pocket, the oxyanion hole-forming loop is disorganized and not functional while residue Leu345 occupies the cavity as a gate occluding the entrance. (B) The L2 loop adopts an intermediate conformation, where the side chain of the residues Leu345 and Lys346 are not opposed but parallel, while the rest of the elements involved in the catalytic mechanism remains similar to the inactive state A. (C) Leu345 and Lys346 have evolved to an active position from the disorganized intermediate B, inverting the orientation of their side chains originally displayed in their active conformations. This movement unblocks the S1 site of the protease while the remaining elements (catalytic triad, oxyanion hole forming loop and L3 loop) are in an inactive-like state as in state A and B. (D) The catalytic triad and all the remaining elements are now aligned in an active configuration. Only the oxyanion hole-forming loop remains disordered and not functional. (E) The structure is completely active and resembles the crystallographic active conformation (PDB 3NZI). Loop L3 stabilizes to adopt a well-defined secondary structure.