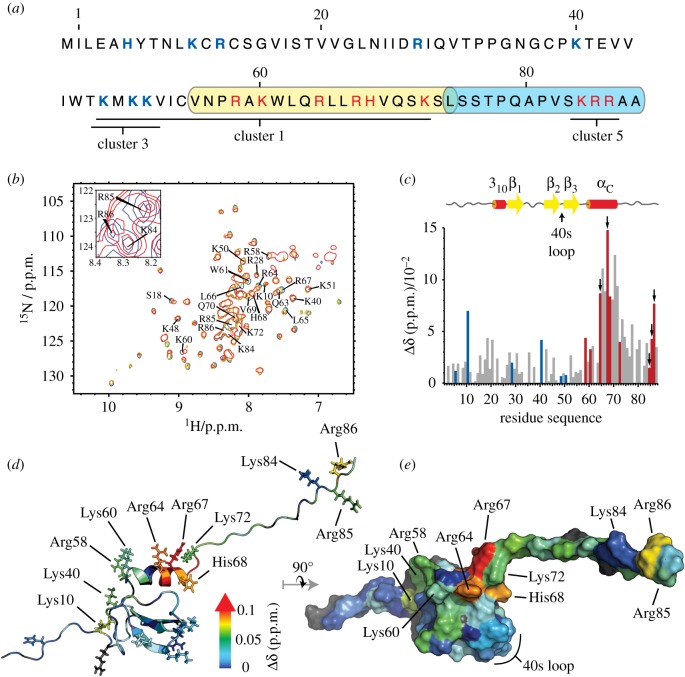
Figure 3.
CXCL13-HS binding sites. (a) CXCL13 sequence highlighting the three putative HS binding clusters 1, 3 and 5 (underlined). The coverage of CXCL13 sequence by the two heparin-covalently bound peptides (yellow and blue boxes) includes nine basic residues (in red) and matches the above-mentioned predicted clusters 1 and 5 binding sites. (b) The [15N,1H]-correlation spectra recorded on Met-CXCL13 at 37 µM alone (blue) or in presence of 37 µM (green), 74 µM (orange) or 144 µM (red) of dp4 have been overlaid to highlight the CSPs of CXCL13 amides upon binding to HS. The inset displays the spectra recorded on sample without (blue) or with 144 µM of dp4 (red) to highlight the CSPs experienced by the C-terminal basic cluster. (c) The combined CSPs of CXCL13 (37 µM) amides upon binding to HS dp4 (74 µM) were plotted against the amino acid sequence number. Coloured CSPs correspond to the blue and red residues in (a). Basic residues experiencing large CSPs, which are thus likely to directly interact with HS (arrows), were selected as candidates for point mutations. (d,e) Cartoon or surface representation of CXCL13 structure, on which CSP values are mapped using a blue to red colour gradient from 0 to a specified threshold, above which the colour was set red. The most affected residues are localized within the α-helix and C-terminal flexible tail. In opposition, the 40s-loop-included residues (highlighted in e) remained unaffected upon dp4 binding.