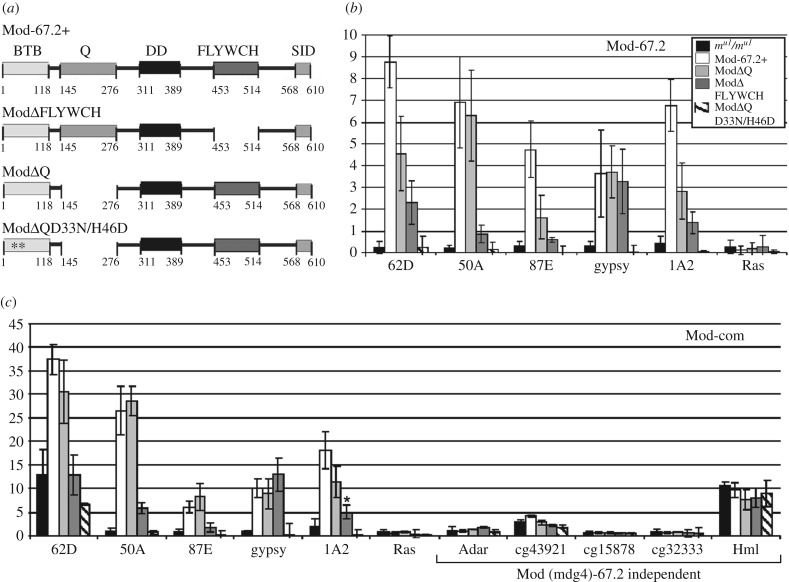
Figure 7.
Role of FLYWCH and Q-rich domains in recruiting Mod(mdg4)-67.2 to chromatin in vivo. (a) Schemes of Mod(mdg4)-67.2 and its deletion derivatives: Mod-67.2+ is the wild-type protein, ModΔFLYWCH lacks the FLYWCH domain, ModΔQ lacks the Q-rich domain, and ModΔQD33N/H46D is a double mutant that lacks the Q-rich domain and has the most conserved aspartate (33) and histidine (46) in the BTB domain substituted by asparagine and acidic aspartate, respectively (indicated with asterisks). Other designations are as in figure 2a. (b,c) ChIP-qPCR analysis of Mod-67.2, and Mod-com binding in middle pupae of the above transgenic lines. PCR products were amplified from two separate immunoprecipitates of three different chromatin preparations. Error bars indicate the standard deviation of three independent biological replicates. *p ≤ 0.05 (Student's t-test), in other cases, p ≤ 0.01. The ras64B coding region (Ras) was used as a control devoid of Su(Hw)-binding sites. Other designations are as in figure 1b. Analysis of transgenic lines was performed in the y2scD1ct6; P{Act5C-GAL4}25FO1/+; mod(mdg4)u1/mod(mdg4)u1 background.