ABSTRACT
Campylobacter jejuni is a foodborne pathogen that is recognized as the leading cause of human bacterial gastroenteritis. The widespread use of antibiotics in medicine and in animal husbandry has led to an increased incidence of antibiotic resistance in Campylobacter. In addition to a role in multidrug resistance (MDR), the Campylobacter CmeABC resistance-nodulation-division (RND)-type efflux pump may be involved in virulence. As a vehicle for pathogenic microorganisms, the protozoan Acanthamoeba is a good model for investigations of bacterial survival in the environment and the molecular mechanisms of pathogenicity. The interaction between C. jejuni 81-176 and Acanthamoeba polyphaga was investigated in this study by using a modified gentamicin protection assay. In addition, a possible role for the CmeABC MDR pump in this interaction was explored. Here we report that this MDR pump is beneficial for the intracellular survival and multiplication of C. jejuni in A. polyphaga but is dispensable for biofilm formation and motility.
IMPORTANCE The endosymbiotic relationship between amoebae and microbial pathogens may contribute to persistence and spreading of the latter in the environment, which has significant implications for human health. In this study, we found that Campylobacter jejuni was able to survive and to multiply inside Acanthamoeba polyphaga; since these microorganisms can coexist in the same environment (e.g., on poultry farms), the latter may increase the risk of infection with Campylobacter. Our data suggest that, in addition to its role in antibiotic resistance, the CmeABC MDR efflux pump plays a role in bacterial survival within amoebae. Furthermore, we demonstrated synergistic effects of the CmeABC MDR efflux pump and TetO on bacterial resistance to tetracycline. Due to its role in both the antibiotic resistance and the virulence of C. jejuni, the CmeABC MDR efflux pump could be considered a good target for the development of antibacterial drugs against this pathogen.
KEYWORDS: Campylobacter jejuni, Acanthamoeba polyphaga, host cell invasion, survival and multiplication, antibiotic resistance, multidrug efflux pumps, biofilm formation, motility, CmeB
INTRODUCTION
Campylobacter jejuni is a microaerophilic, spiral-shaped, Gram-negative, motile, foodborne pathogen that is recognized as the main cause of bacterial gastroenteritis worldwide (1). Most commonly, the disease is associated with consumption of undercooked poultry or contaminated water, where Campylobacter can coexist with protozoa and form biofilms (2). Antimicrobial therapy is warranted for immunocompromised patients or patients with severe infections and, although most people recover quickly from this disease, others may develop rare neurodegenerative disorders such as Guillain-Barré syndrome (GBS), which manifests as paralysis, requiring extensive medical treatment (3).
The ability of C. jejuni to invade host cells is important for pathogenicity (4). To establish infection in humans, C. jejuni invades the gut epithelial layer and colonizes the intestine by employing a variety of virulence factors (2). The CmeABC multidrug resistance (MDR) efflux pump plays a key role in C. jejuni colonization of chickens (a natural host and a major reservoir of Campylobacter) by mediating resistance to bile salts present in the intestinal tract (5). The MDR pump is a tripartite efflux system belonging to the resistance-nodulation-division (RND) superfamily of bacterial transporters. It consists of three components, i.e., the outer membrane channel-forming protein CmeC, the inner membrane drug transporter CmeB, and the periplasmic protein CmeA, which bridges CmeB and CmeC (6). The CmeABC complex contributes to the intrinsic resistance of C. jejuni to a broad range of antibiotics, heavy metals, and other antimicrobial agents (7). There are high levels of variation in the amino acid sequences of the CmeB protein among C. jejuni strains, which may have an impact on the function of this transporter (8). Investigation of the molecular mechanisms of antibiotic resistance is important for control of the dissemination of multidrug-resistant bacteria (9).
Although most studies of MDR pumps have focused on investigations of their role as antibiotic resistance determinants, MDR pumps may also play a role in bacterial pathogenesis (10–15). For example, the CmeB homologues AcrB (Salmonella enterica and Klebsiella pneumoniae) and MexB (Pseudomonas aeruginosa) are required for invasion of host cells and virulence (16–19), suggesting a possible contribution of the CmeB protein to the pathogenic properties of C. jejuni.
The CmeABC efflux pump of C. jejuni is known to be required for resistance to antibiotics, bile salts, and some disinfectants, as well for host colonization (5, 7, 20). However, its role in biofilm formation, motility, or survival within amoebae has not yet been studied.
Acanthamoeba is a genus of amoebae containing free-living protist pathogens that are widely spread in water environments (21, 22). These eukaryotic organisms are characterized by spine-like structures on their surface, which allows them to adhere, to move, and to capture their prey by phagocytosis (21). During their life cycle, acanthamoebae can adopt two reversible forms, i.e., trophozoite cells, which are able to feed on microbes, and dormant, double-walled, cyst cells with minimal metabolic activity, which are formed under adverse environmental conditions such as extreme temperatures or pH values (22). Acanthamoebae are opportunistic pathogens that are capable of causing serious human infections, including blinding keratitis, which is associated mostly with contact lens users, and fatal granulomatous encephalitis, which occurs mainly in immunocompromised patients (21).
An increasing number of microorganisms, such as bacteria and viruses, have been reported to benefit from interactions with these free-living pathogens, as the amoebae play a role as a reservoir, allowing the microorganisms to escape predation and perhaps enabling them to survive and/or to multiply inside their hosts and to be transmitted in the environment (23). Pathogenic microorganisms residing inside amoebae become more virulent and less susceptible to antibiotics and disinfectants, making it difficult to eradicate them from public water supplies and creating problems for human and animal health (24–26).
Amoebae are easy to handle experimentally and represent an attractive and simple model of infection to study host-pathogen interactions in vitro, allowing the discovery of new bacterial virulence factors, which may facilitate the development of new antibacterial therapeutic agents (27). Also, amoebae exhibit features similar to those of macrophages, especially in the way they capture their prey by phagocytosis (28).
Elucidation of the molecular mechanisms involved in these interactions is relevant to public health, since microbial pathogens and acanthamoebae can coexist in the same environments, e.g., in the water of industrial poultry houses (despite stringent biosecurity measures) (29). Such associations may be beneficial to the acanthamoebae and/or the bacteria, which may lead to symbiotic relationships between these microorganisms (30).
The molecular mechanisms of interactions between C. jejuni and these eukaryotic hosts are not well understood. The results from a few publications describing the interactions between Acanthamoeba polyphaga and C. jejuni are contradictory (31). While some studies suggest the ability of C. jejuni to survive and/or to multiply within amoebae (32–39), others support an extracellular mode of survival only (40–43). These conflicting results may be explained by variations in the strains of C. jejuni and amoebae and the use of different methods, such as whether the in vitro survival experiments involved gentamicin treatment.
The objectives of this study were (i) to elucidate the type of interactions between C. jejuni and A. polyphaga, (ii) to investigate whether the CmeB multidrug efflux transporter is involved in this interaction, (iii) to explore other possible biological functions for this transporter in C. jejuni, and (iv) to investigate the contribution of variations in the amino acid sequence encoded by cmeB to the tetracycline resistance of C. jejuni. To accomplish the first two aims, the standard gentamicin protection method was modified to ensure more efficient elimination of both attached and extracellular bacteria (which could escape to the medium and cause reinfection, thus potentially producing misleading data).
The association between foodborne pathogens and protozoa leads to serious consequences for food safety, increasing the risk of infection (44). Deciphering the molecular mechanisms of Campylobacter-amoeba interactions will facilitate a better understanding of the lifestyle of this foodborne pathogen, aiding in the development of novel intervention strategies.
RESULTS
C. jejuni 81-176 is able to survive and to multiply within A. polyphaga.
The survival of C. jejuni at different temperatures was investigated in vitro by using the A. polyphaga model of infection. Due to its lifestyle, Campylobacter is likely to encounter a wide range of environmental conditions, including different temperatures. In order to survive, it must be able to sense, to adapt to, and to respond to temperature fluctuations (45). The temperatures 25°C and 37°C were selected for this study, to mimic environmental and human host temperatures, respectively.
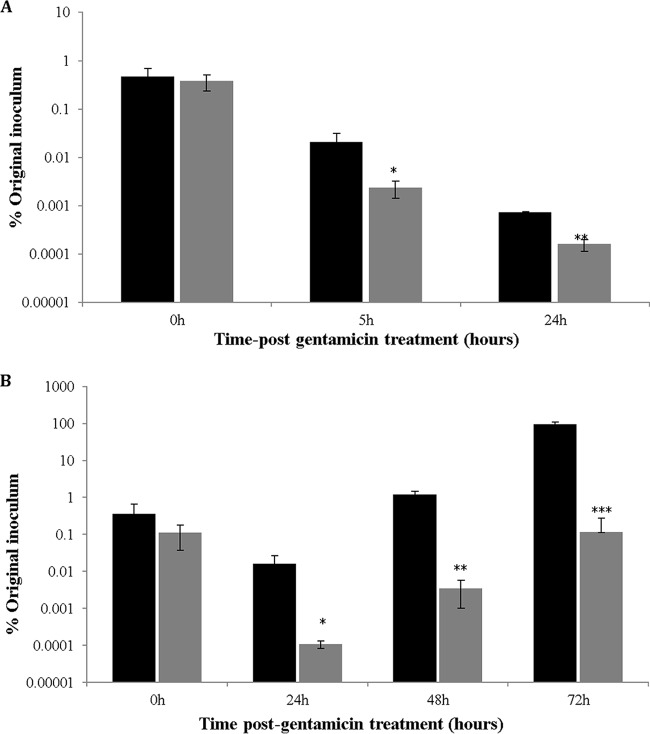
To elucidate the interaction between C. jejuni 81-176 and A. polyphaga, the standard gentamicin protection method (19) was modified by adding an extra 1 h of gentamicin treatment at the later incubation time points, before lysis of the cells; this avoided quantification of bacteria that were attached or had escaped to the extracellular medium and that were potentially capable of reinvasion, since the gentamicin concentration used (100 μg/ml) was sufficient to kill C. jejuni wild-type (WT) and mutant strains (data not shown). By using the modified version of this method, we observed a substantial decrease in the numbers of intracellular bacteria, compared with the standard version, at both 25°C (Fig. 1A) and 37°C (Fig. 1B).
FIG 1.
C. jejuni 81-176 is able to survive and to multiply inside A. polyphaga. (A) Intracellular survival was determined by CFU counting at 0, 5, and 24 h after gentamicin treatment at 25°C, under aerobic conditions. (B) Intracellular multiplication was determined at 0, 24, 48, and 72 h after gentamicin treatment at 37°C, under aerobic conditions. Black bars represent bacterial counts obtained with the standard gentamicin protection assay, and gray bars represent bacterial counts obtained with a modified version developed in this study. P values, referring to comparisons between the samples at each time point, were as follows: panel A: 0 h, P = 0.583; 5 h, P = 0.031; 24 h, P = 0.00003; panel B: 0 h, P = 0.166; 24 h, P = 0.03; 48 h, P = 0.001; 72 h, P = 0.00028. *, 0.01 < P ≤ 0.05; **, 0.001 < P ≤ 0.01; ***, P ≤ 0.001.
At 25°C and 0 h (defined as the time point immediately after the first gentamicin treatment), the methods produced similar CFU counts (Fig. 1A). However, much greater decreases in intracellular bacteria were observed at 5 h and 24 h with the modified procedure (Fig. 1A). Since no colonies were detectable at this temperature after 48 h (data not shown), no further time points are shown. These data indicate that strain 81-176 can invade and survive inside amoebae at 25°C for a certain period.
Because the optimal growth temperature for C. jejuni is 37°C, time points for this temperature were extended to 72 h. At this temperature, the decrease in the intracellular bacterial numbers at 24 h after gentamicin treatment with the modified gentamicin method was even more pronounced, and even greater differences were observed at the later time points (Fig. 1B). With both methods, initial reductions in the CFU counts were followed by increases after prolonged incubation, suggesting bacterial multiplication (Fig. 1B). Although the methods demonstrated the same trends in changes in CFU counts with time, the modified version allowed more accurate quantification of only intracellular bacteria and was used in all subsequent experiments.
A different modification of the gentamicin protection method, in which a lower concentration of gentamicin is constantly maintained, was described elsewhere (46). Using that method, however, we observed that C. jejuni 81-176 was not able to multiply intracellularly and was not detected 48 h postinfection (data not shown), probably because the antibiotic was able to enter the amoebic cells during prolonged incubations. This observation is in accordance with previous studies reporting the ability of gentamicin to enter host cells during prolonged incubations and to kill intracellular bacteria (47, 48).
The numbers of viable extracellular bacteria were monitored for a 6-day period. At 96 h postinfection, a significant increase in bacterial counts was observed, compared with bacteria incubated in medium alone. Moreover, after 6 days of incubation, the presence of amoebae allowed the isolation of viable bacteria, while none could be detected in the absence of amoebae (see Fig. S1 in the supplemental material). In summary, these data support both extracellular and intracellular modes of survival for C. jejuni 81-176 in cocultures with A. polyphaga at different temperatures.
CmeB contributes to C. jejuni 81-176 intracellular survival and multiplication within A. polyphaga.
To check whether the C. jejuni CmeB transporter is required for survival and multiplication within amoebae, the cmeB gene of strain 81-176 was inactivated by insertional mutagenesis (49) to create the 81-176/cmeB::kanr mutant. Complementation was achieved by replacement of the mutated gene with its wild-type copy and selection of the derivative at a tetracycline concentration that did not support the growth of the mutant strain (Table 1). Construction of the mutant strain and its complementation derivative was confirmed by PCR (Fig. S2A). The mutation had no impact on the bacterial growth rate (Fig. S2B).
TABLE 1.
Susceptibility of C. jejuni WT strains and their cmeB mutants to tetracycline
| Campylobacter jejuni strain | Tetracycline MIC (mean ± SD) (μg/μl)a | Fold change (WT strain vs mutant strain) |
|---|---|---|
| 81-176/cmeB+/pTet+ (WT) | 62.5 ± 0.010 | |
| 81-176/cmeB−/pTet+ | 7.8 ± 0.020 | 8 |
| 11168H/cmeB+/pTet− (WT) | 0.12 ± 0.017 | |
| 11168/cmeB−/pTet− | 0.06 ± 0.017 | 2 |
| G1/cmeB+/pTet− (WT) | 1.95 ± 0.002 | |
| G1/cmeB−/pTet− | 0.06 ± 0.011 | 32.5 |
| G1/cmeB+/pTet+ | 500 ± 0.031 | 256 |
MICs for tetracycline were determined by the microdilution method in MH-F broth, according to the EUCAST recommendations (64). Tetracycline MICs were determined for strains 81-176, 11168H, and G1 and their respective cmeB mutants; three clonal isolates were tested for each strain. The tetracycline MIC value was also determined for a derivative of the G1 strain containing the pTet plasmid from C. jejuni strain 81-176.
The tissue culture experiments involved lysis of the amoebic cells with 0.1% Triton X-100 for 15 min at room temperature. Because the 81-176 cmeB mutant strain was shown to be susceptible to this detergent (5), experiments were performed to ensure that the data obtained were genuine and not experimental artifacts. The effect of 0.1% (vol/vol) Triton X-100 was investigated by simulating its use in cell culture experiments in which this detergent was added to the bacteria for a short period. Under those conditions, CFU counts for the 81-176/cmeB::kanr mutant were similar to those of the WT and complemented strains (Fig. S3), suggesting that Triton X-100 had no detrimental effect on bacterial viability.
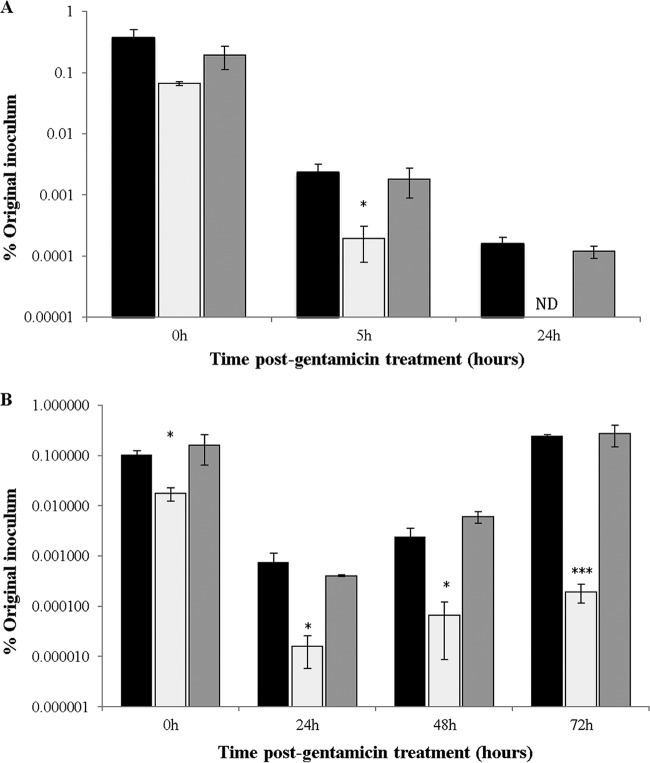
At both 25°C and 37°C, the numbers of intracellular bacteria for the 81-176/cmeB::kanr mutant were significantly lower than those of the wild-type strain, with the difference increasing after longer incubations (Fig. 2A). In contrast to the WT strain, no amplification of bacteria was observed at 37°C even after 72 h (Fig. 2B). There was no statistically valid difference in CFU numbers for the mutant at 48 h and 72 h, compared to those at 24 h (P = 0.24 and P = 0.09, respectively). Complementation of the 81-176 cmeB mutant strain restored the phenotype in all experiments. In summary, the results suggest that the C. jejuni 81-176 CmeB is required for bacterial survival and multiplication in acanthamoebae.
FIG 2.
CmeB is required for survival and multiplication of C. jejuni 81-176 in A. polyphaga. (A) Intracellular survival was determined by CFU counting at 0, 5, and 24 h after gentamicin treatment at 25°C, under aerobic conditions. (B) Intracellular multiplication was determined at 0, 24, 48, and 72 h after gentamicin treatment at 37°C, under aerobic conditions. Black bars, 81-176; white bars, 81-176/cmeB::kanr mutant; gray bars, 81-176/cmeB::kanr/cmeB complementation derivative. P values, referring to comparisons between the values for WT and mutant strains at each time point, were as follows: panel A: 0 h, P = 0.018; 5 h, P = 0.015; panel B: 0 h, P = 0.021; 24 h, P = 0.031; 48 h, P = 0.028; 72 h, P = 0.000004. *, 0.01 < P ≤ 0.05; ***, P ≤ 0.001. ND, not detected.
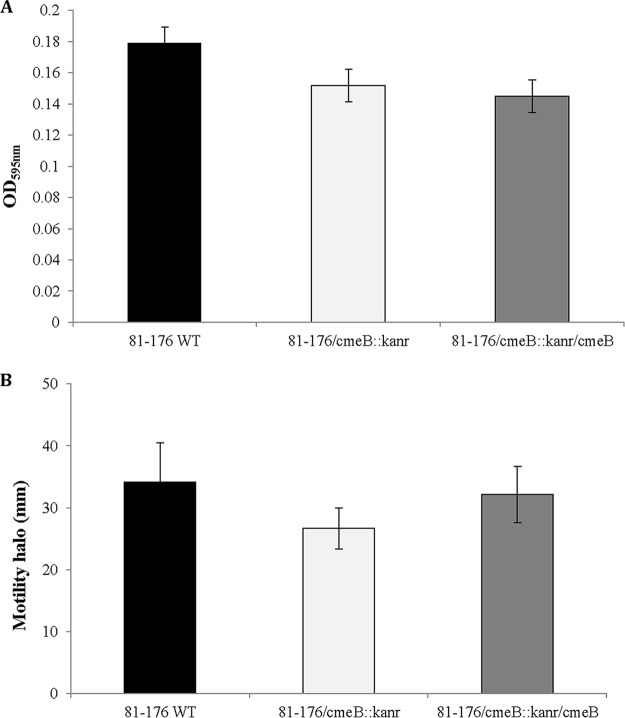
CmeB is not required for biofilm formation and motility of C. jejuni 81-176.
The ability to form biofilms is an important factor in the lifestyle of C. jejuni (50). Some studies showed that inactivation of the multidrug efflux pumps could prevent biofilm formation (51). Using a previously described assay to study pellicle formation at the air-liquid interface (52), we found no effect of the cmeB mutation on the ability of the bacteria to form this type of biofilm (Fig. 3A). Since C. jejuni motility plays an important role in the invasion of host cells (53), we also aimed to verify whether mutated bacteria remained motile. As shown in Fig. 3B, the cmeB mutation did not have any effect on bacterial motility.
FIG 3.
The cmeB mutation does not affect biofilm formation and motility. (A) Quantification of biofilms at the air-liquid interface of the glass tubes. The absorbance values measured for the WT, cmeB mutant, and complemented strains were 0.179 ± 0.06, 0.151 ± 0.01, and 0.145 ± 0.02, respectively. No statistically significant difference (P = 0.443) in biofilm quantities between the WT and cmeB mutant strains was observed. (B) Quantification of growth zones in BHI soft-agar motility plates inoculated with different C. jejuni strains. The average diameters of bacterial growth for the WT, cmeB mutant, and complemented strains were 34.2 ± 6.37 mm, 26.7 ± 3.33 mm, and 32.2 ± 4.54 mm, respectively. No statistically significant difference in growth zones (P = 0.15) between the WT and cmeB mutant strains was observed. Black bars, 81-176; white bars, 81-176/cmeB::kanr mutant; gray bars, 81-176/cmeB::kanr/cmeB complementation derivative.
The CmeABC transporter and sequence variations of the CmeB protein contribute to tetracycline resistance.
The tetracycline resistance of some Campylobacter strains is associated with the tetO gene carried by a pTet plasmid (6). In a previous study, we observed that C. jejuni strain G1, which was isolated from a patient with GBS, was more resistant to tetracycline than was the 11168H strain, despite the absence of the tetO gene in both strains (54). Comparison of the genome of the G1 strain with that of reference strain NCTC 11168 revealed a remarkable difference in the sequences of the cmeB genes (54). The protein sequence identity for cmeB genes from these two strains was only 81%, while that for genes from strains 11168H and 81-176 was as high as 99%. In this study, we found that inactivation of cmeB in the G1 strain resulted in greater reduction of resistance to tetracycline than did inactivation of cmeB in the 11168H strain (Table 1). However, transfer of the pTet plasmid from C. jejuni 81-176 to the G1 strain by conjugation made the latter 8 times more resistant to tetracycline than the donor C. jejuni 81-176 carrying this plasmid (Table 1). This finding suggests that the CmeABC MDR efflux pump of C. jejuni strain G1 has greater ability to excrete this drug than do those of C. jejuni strains 11168H and 81-176. Since the nucleotide sequence differences were predominantly limited to cmeB, the variation in the efficiency of these pumps may be primarily associated with the product of this gene (Tables S1 and S2).
In addition, it was found that C. jejuni strain G1 was more resistant than strain 81-176 to various antibiotics and that the disruption of cmeB (confirmed with three clonal isolates) increased the susceptibility of the G1 strain to almost all antibiotics tested. The exceptions were penicillin, linezolid, and trimethoprim, for which no difference could be found due to very low (undetectable) levels of inhibition, probably due to low concentrations of the antibiotics used (Fig. S4). These data show that the spectrum and level of antimicrobial resistance of C. jejuni may be primarily dependent on the sequence variations of cmeB, with the potential for certain mutations in this gene to result in “super” efflux pump variants, significantly enhancing bacterial resistance to multiple antibiotics.
DISCUSSION
The association between free-living amoebae and pathogenic bacteria is concerning, because it may have significant implications for human health (55). In this study, the interaction between C. jejuni 81-176 and A. polyphaga was investigated. C. jejuni displays extensive genetic variation; consequently, the efficiency by which C. jejuni interacts with cultured cells depends on specific properties of the strains (2, 56).
This study suggests that, during longer periods of incubation with A. polyphaga at 37°C, intracellular C. jejuni may escape into the extracellular medium, multiply, and reinfect amoebic cells. As the result, reinfection rather than intracellular multiplication may be the primary factor responsible for increased intracellular numbers. In order to obtain more accurate data about intracellular multiplication, an additional gentamicin treatment step was introduced in this study. Compared with the standard gentamicin assay, much more significant reductions in intracellular bacterial numbers were observed at both 25°C and 37°C. In the future, it would be interesting to investigate the interaction between C. jejuni and A. polyphaga at 42°C, as this is the chicken body temperature. It was observed previously that, at that temperature, the amoebic cells changed to the cyst shape (21). This is relevant, since amoebic cysts may play a role in contamination and the persistence of pathogenic bacteria in food-related environments, allowing internalized foodborne pathogens to resist the disinfection treatments used in the food industry (57). Despite gradual decreases in the numbers of intracellular bacteria at 25°C, the fact that intracellular bacteria are still detectable after 24 h might be of epidemiological importance if the amoebae are constantly exposed to the presence of these bacteria in the environment.
Our data also demonstrated prolonged extracellular survival of strain 81-176 in the presence of amoebae, which is in accordance with previously published results (41). According to Bui et al., this is likely to be due to the depletion of dissolved oxygen by amoebae, thus creating the microaerophilic environment optimal for C. jejuni growth (41).
At 37°C, the initial decrease in the number of viable bacteria was followed by a remarkable increase after 48 h of incubation. A similar trend was reported for S. enterica and Listeria monocytogenes (58). The authors termed the initial decrease in viability the eclipse phase, which probably occurs due to the initial use of the bacteria as a food source or due to a prolonged lag or adaptation phase, followed by active intracellular growth (58).
Although the CmeABC efflux pump of C. jejuni was reported previously to be required for colonization of the intestinal tract of chickens (5), its role in bacterium-host cell interactions was not determined. We were able to show that this pump is beneficial for the survival and replication of C. jejuni 81-176 within amoebae and that the reduction in the CFU numbers for the cmeB mutant was not the result of its greater sensitivity to the detergent used for amoebic lysis. These observations support the idea that efflux pumps may indeed act as virulence determinants. The role of MDR efflux pumps in virulence could be linked to their ability to expel and to confer resistance to host-derived antimicrobial agents, such as low-molecular-weight toxins (reactive oxygen species) and antimicrobial peptides (14).
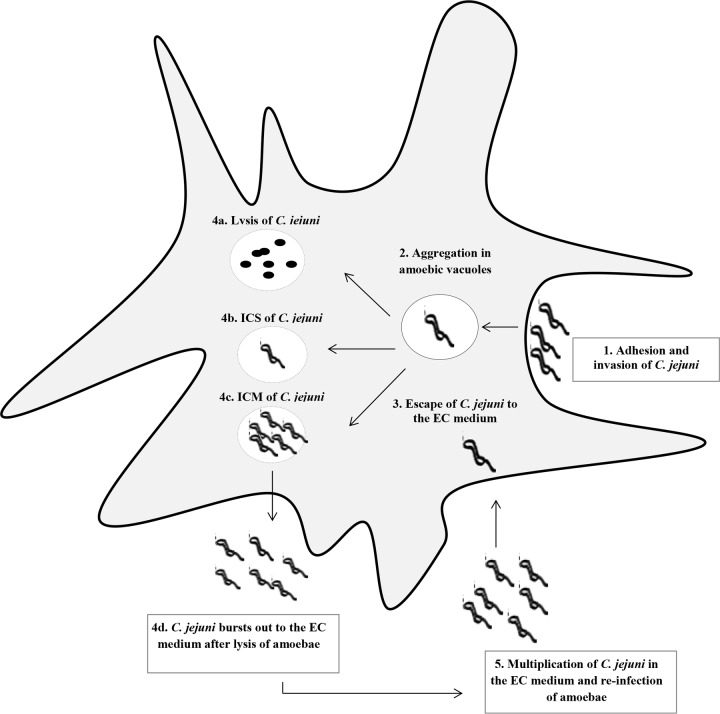
Based on the information available in the literature and the data presented here, we suggest a hypothetical model describing a mechanism of interaction between C. jejuni and the amoebae (Fig. 4). According to this model, intracellular bacteria acquired from the environment (e.g., at 25°C, as in our experiments) multiply at 37°C (conditions simulating the host temperature). After ingestion of a product (e.g., water or milk) contaminated with amoebae, the latter are lysed, releasing large amounts of bacteria causing disease. A global search for other bacterial factors involved in the interaction between C. jejuni and amoebae could be based on differential expression studies (transcriptomics and proteomics). Our data also suggested that neither motility nor biofilm formation was responsible for the decreased survival of the cmeB mutant strain within the A. polyphaga host.
FIG 4.
Hypothetical model of the interaction between C. jejuni and A. polyphaga. The following possible stages of bacterial entry are depicted: 1, adhesion to and invasion of amoebic cells via phagocytosis; 2, gathering within amoebic vacuoles (38, 40); 3, escape to the extracellular (EC) medium (39); 4a, bacterial cell lysis; 4b, intracellular survival (ICS) and escape without lysis; 4c, intracellular multiplication (ICM); 4d, escape after lysis followed by release into the extracellular medium (39); 5, presence in the extracellular medium. In the extracellular medium, C. jejuni is able to multiply and to reinfect other amoebic cells. Stages 1, 3, 4a to 4c, and 5 are based on the observations reported in this study.
As demonstrated in this study, the exchange of genetic elements involved in antibiotic resistance can result in dramatic elevation of C. jejuni resistance to antibiotics. This is in accordance with other studies reporting C. jejuni CmeB sequence variants with much more powerful efflux of antibiotics, leading to enhanced antibiotic resistance (8, 59). The CmeB from C. jejuni strain G1 in this study shared 99% identity with the two super efflux pump variants discovered previously, i.e., the 154KU variant (8) and the RE-CmeB variant (59), suggesting that the CmeB from the G1 strain might also be a super efflux variant. However, this hypothesis needs to be supported by further studies. Despite much greater strain-to-strain sequence variation of CmeB, compared with other components of the MDR pump and CmeR, further experiments are required for evaluation of the relative contributions of these proteins to variations in the levels of antibiotic resistance. In this study, it was possible to generate bacteria with tetracycline resistance levels remarkably exceeding those of all parental strains. Such exchanges are likely to occur in the environment not only via conjugation (e.g., involving a transfer of the pTet plasmid) but also via transformation, as many strains of C. jejuni are naturally competent and can easily acquire DNA released due to the lysis of cells carrying antibiotic resistance genes (2). As a result, originally sensitive C. jejuni strains might easily become more resistant to antibiotics. Variation in CmeB structure is just one of various strategies utilized by bacteria for adaptation to hostile environmental conditions, both in vivo and ex vivo.
To summarize, we describe here the contribution of a Campylobacter efflux pump to bacterial survival within amoebae, which was demonstrated by employing an improved gentamicin protection method. Moreover, using a modified version of the gentamicin treatment method, the results explain the previously reported controversial results from investigations of Campylobacter-amoeba interactions. Because CmeABC is required for C. jejuni antibiotic resistance and virulence, this efflux pump is a promising target for interventions to combat C. jejuni infections.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni strain 81-176 was used throughout this study because of its high virulence (60, 61) and enhanced ability to invade and to survive within A. polyphaga (according to our data). C. jejuni 11168H, a hypermotile variant of the reference strain NCTC 11168 (62), and G1, a strain isolated from a patient with Guillain-Barré syndrome (63), were used for the tetracycline resistance assay. Strain X was isolated from a patient with enteritis (49). C. jejuni strains were routinely grown for 24 h at 37°C on Columbia blood agar (CBA) (Oxoid) supplemented with 5% defibrinated horse blood and Campylobacter selective supplement (Skirrow; Oxoid), under microaerobic conditions (5% O2 and 10% CO2 in N2). NEB Express competent Escherichia coli used for molecular cloning was grown at 37°C in Luria-Bertani (LB) medium (Oxoid). When appropriate, the following antibiotics were included in the culture medium as selective agents: ampicillin (100 μg/ml), kanamycin (50 μg/ml), and tetracycline (10 μg/ml).
For liquid cultures, C. jejuni was suspended in brain heart infusion (BHI) broth (Oxoid) and adjusted to an optical density at 600 nm (OD600) of 1. The bacterial suspension was diluted 100-fold in BHI broth, in sterile conical flasks, and was incubated microaerobically at 37°C for 2 days, with shaking at 200 rpm. One-milliliter samples of each bacterial culture were taken at each time point (0, 6, 24, 30, and 48 h), and the OD600 was measured.
Generation of C. jejuni 81-176 cmeB mutant and complemented strains.
Inactivation of the C. jejuni 81-176 gene cmeB was achieved by an insertional mutagenesis approach (49), in which cmeB was disrupted by insertion of a kanamycin resistance (kanr) cassette. The primers CmeB-F (5′-AAGGAGATATACCATGTTTTCTAAATTTTTTATAGAAAGACCTATTTTTG-3′) and CmeB-R (5′-TCATTCATGAATCTTACCTCTTTTTTTATCTAGC-3′) were used to amplify a 3-kb fragment containing the cmeB gene from C. jejuni 81-176 chromosomal DNA. The PCR product was ligated with pGEM-T Easy vector (Promega) using T4 DNA ligase (New England BioLabs), followed by transformation into NEB Express competent E. coli and selection of recombinant clones. The kanr cassette was isolated from vector pJMK30 by digestion with SmaI (1.5 kb), followed by gel extraction of the 1.5-kb fragment containing the kanr cassette. The plasmid pGEM-T Easy/cmeB was digested with ClaI and ligated with a DNA fragment containing the kanr cassette by using T4 DNA ligase (Promega). After transformation of E. coli with the ligation mixture, the pGEM-T Easy/cmeB::kanr plasmid containing the insert of interest, in the correct orientation, was isolated. The pGEM-T Easy/cmeB::kanr plasmid was transformed into C. jejuni 81-176 via electroporation, and transformants were selected on CBA supplemented with kanamycin (50 μg/ml). The C. jejuni 81-176/cmeB::kanr mutants were confirmed by PCR analysis.
The complemented strain C. jejuni 81-176/cmeB::kanr/cmeB was constructed via homologous recombination. The genomic DNA from the C. jejuni 81-176 strain was transformed into competent cells of the cmeB mutant strain via electroporation, and transformants were selected in CBA supplemented with tetracycline at 20 μg/ml, a concentration that does not support the growth of the cmeB mutant strain. A similar procedure was used for the construction of complementation derivatives of other strains (Table 1).
Antibiotic susceptibility assays. (i) Broth microdilution assay.
The MICs were determined according to EUCAST guidelines (64). The MICs of tetracycline (Sigma) for C. jejuni strains were determined by a broth microdilution method, using Mueller-Hinton-fastidious (MH-F) broth (cation-adjusted Mueller-Hinton [MH] broth with 5% lysed blood and 20 mg/liter β-NAD). Briefly, 10 μl of C. jejuni suspension in MH-F broth (OD600 of 0.5) was added to 90 μl of 2-fold dilutions of tetracycline in MH-F broth. Suspensions were transferred to a 96-well flat-bottomed microtiter plate (Corning) and incubated for 3 days at 37°C under microaerobic conditions, with shaking at 100 rpm. The tetracycline concentration range tested was 0.03 μg/ml to 500 μg/ml, and control wells with no tetracycline were included. MICs were measured at 600 nm using an Infinite 200 PRO plate reader (Tecan). According to EUCAST guidelines, the tetracycline MIC breakpoints for C. jejuni are as follows: sensitive, ≤2 μg/μl; resistant, >2 μg/μl (64).
(ii) Antibiotic disc diffusion assay.
The antibiotic susceptibility of C. jejuni strains was determined according to the EUCAST recommendations (64). C. jejuni was grown on CBA plates for 24 h at 37°C and suspended in 1 ml of MH broth at an OD600 of 0.5. The suspension (100 μl) was spread on MH agar plates supplemented with 5% lysed horse blood, using a spreader. Antibiotic-containing discs (Oxoid) were placed at the surface of the agar plates using sterile tweezers, and the plates were incubated for 2 days at 37°C under microaerobic conditions. Inhibition zone diameters were measured and interpreted accordingly to EUCAST zone diameter breakpoints for 30-μg tetracycline discs, as follows: sensitive, ≥30 mm; resistant, <30 mm (64).
Amoebic culture conditions.
Acanthamoeba polyphaga (Linc Ap-1), which was used in all experiments, was kindly provided by Bernard de La Scola, University de La Mediterranee (Marseille, France). A. polyphaga was maintained aerobically at 25°C in peptone-yeast-glucose (PYG) medium (20 g protease peptone, 18 g glucose, 1 g yeast extract, 1 g MgSO4·7H2O, 1 g sodium citrate·2H2O, 0.02 g Fe(NH4)2(SO4)2·6H2O, 0.06 g CaCl2, 0.14 g H2PO4, and 0.35 g Na2HPO4·7H2O in 1 liter [pH 6.8]; filter sterilized with a 0.22-μm filter), in 75-cm2 treated culture flasks. The concentration and viability of amoebae were determined by the trypan blue exclusion assay, and amoebae were visualized by phase-contrast microscopy with a ×40 objective in an inverted cell culture microscope (Motic AE31).
To test the sensitivity of the C. jejuni 81-176 cmeB mutant to 0.1% (vol/vol) Triton X-100, bacterial cells were harvested from overnight CBA plates, suspended in BHI medium, and adjusted to an OD600 of 1 (initial inoculum). As controls, 100-μl aliquots of the initial inocula were serially diluted in phosphate-buffered saline (PBS) and plated on CBA plates for CFU counting. The bacterial suspensions were then centrifuged for 2 min at 10,000 × g, 1 ml of 0.1% (vol/vol) Triton X-100 was added, and the cells were incubated for 15 min at room temperature. Bacterial suspensions in Triton X-100 detergent were serially diluted in PBS and plated on CBA plates. The CFU counts obtained for the mutant strain were compared with those obtained for the WT and complemented strains.
In vitro coculture assay.
Coculturing of C. jejuni with attached monolayers of A. polyphaga cells was conducted in 24-well plates. A. polyphaga cells were seeded at a density of 106 amoebae per ml in PYG medium and were incubated at 25°C for 2 h to allow the cells to settle and to form monolayers at the bottom of the wells. Bacterial cells were harvested from overnight CBA plates, suspended in PYG medium, and adjusted to an OD600 of 1. Then, 100 μl of bacterial suspension was added to the wells with A. polyphaga, achieving multiplicities of infection ranging from 100 to 400 bacteria per well. To allow invasion to occur, cocultures were incubated under aerobic conditions for 2 h at 25°C or 37°C. Following coincubation, wells were washed once with PYG medium and treated for 1 h with 100 μg/ml gentamicin; this concentration was shown to be effective in fully eliminating the bacterial cells in 1 h (data not shown). Following gentamicin treatment, the wells were washed three times with PBS, and the amoebae were lysed with 0.1% (vol/vol) Triton X-100 for 15 min at room temperature, releasing the intracellular C. jejuni. Samples were serially diluted in PBS and plated on CBA plates, in duplicate, followed by 2 days of incubation at 37°C under microaerobic conditions. For longer incubations (24, 48, and 72 h), the wells were incubated with PYG medium without gentamicin. At the designated time points, prior to the addition of 0.1% Triton X-100, the wells were retreated with gentamicin (100 μg/ml) for 1 h (in a modification of the standard gentamicin protection assay). Cells were washed three additional times with PBS, lysed, diluted, and plated as described above. To calculate the number of intracellular bacteria, the following formula was used: [(recovered C. jejuni [in CFU per milliliter])/(total C. jejuni [in CFU per milliliter])] × 100 = % intracellular C. jejuni in A. polyphaga.
Biofilm formation assay.
C. jejuni was grown on CBA plates at 37°C for 24 h, suspended in BHI broth, and adjusted to an OD600 of 0.5. The bacterial suspension (1 ml) was transferred to borosilicate glass tubes and incubated statically at 37°C for 4 days under microaerobic conditions, after which an attached pellicle at the gas-liquid interface was observed (52). For crystal violet (CV) staining, the glass tubes were washed twice with distilled water and dried at 85°C for 30 min. Then, 0.5% CV solution was added to the tubes, and the tubes were incubated at room temperature for 30 min, with gentle shaking. The tubes were washed with distilled water and dried at 85°C for 30 min. Lastly, 1 ml of a 80% ethanol-20% acetone mixture was added for 15 min, to dissolve the CV stain. Samples were transferred to 96-well plates in triplicate, and the OD595 was measured using an Infinite 200 PRO plate reader (Tecan).
Motility assay.
The motility of C. jejuni was determined as described previously, with a few modifications (65). C. jejuni bacteria were grown on CBA plates at 37°C for 24 h, suspended in BHI broth, and adjusted to an OD600 of 0.5, after which 1-μl aliquots of the bacterial suspension were spotted onto 0.4% BHI soft agar plates. The low density of the agar allowed the bacteria to move within the agar, forming a halo of growth around the point of inoculation. Plates were incubated for 3 days at 37°C under microaerobic conditions.
Conjugation of C. jejuni.
To generate the C. jejuni G1/pTet strain, the pTet plasmid from C. jejuni 81-176 was transferred to C. jejuni G1 by conjugation. A mixture of 200 μl of the 81-176 donor strain with 100 μl of the G1 recipient strain (MH medium inoculum at an OD600 of 1) was plated on top of a 0.22-μm filter membrane (Millipore), placed on the surface of a CBA plate, and incubated overnight at 37°C under microaerobic conditions. The bacterial growth was scraped from the membrane and plated on CBA supplemented with tetracycline (15 μg/ml) and ampicillin (5 μg/ml); the latter does not support the growth of the 81-176 strain but allows the G1 strain to grow. PCR was conducted to confirm the derivative strain using the primers pTet-F (5′-GGCGTTTTGTTTATGTGCG-3′) and pTet-R (5′-ATGGACAACCCGACAGAAGC-3′).
Statistical analysis.
All experiments were repeated three times (biological replicates), with three technical replicates in each experiment, and the data were expressed as mean ± standard deviation (SD). Comparisons of two groups were made with an unpaired, two-tailed, Student's t test. Mean differences were considered statistically nonsignificant when the P values were >0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bernard de la Scola for kindly providing us with the Acanthamoeba polyphaga strain.
This work was funded by the Faculty of Science, Engineering, and Computing, Kingston University, through a full-time research degree studentship awarded to A.V.
No potential conflicts of interest were disclosed.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01600-17.
REFERENCES
- 1.Johnson TJ, Shank JM, Johnson JG. 2017. Current and potential treatments for reducing Campylobacter colonization in animal hosts and disease in humans. Front Microbiol 8:487. doi: 10.3389/fmicb.2017.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young KT, Davis LM, DiRita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 3.Wijdicks EFM, Klein CJ. 2017. Guillain-Barré syndrome. Mayo Clin Proc 92:467–479. doi: 10.1016/j.mayocp.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. 2010. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol 300:205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Sahin O, Michel L, Zhang O. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iovine NM. 2013. Resistance mechanisms in Campylobacter jejuni. Virulence 4:230–240. doi: 10.4161/viru.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J, Michel L, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cagliero C, Cloix L, Cloeckaert A, Payot S. 2006. High genetic variation in the multidrug transporter cmeB gene in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 58:168–172. doi: 10.1093/jac/dkl212. [DOI] [PubMed] [Google Scholar]
- 9.Bolton DJ. 2015. Campylobacter virulence and survival factors. Food Microbiol 48:99–108. doi: 10.1016/j.fm.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Blanco P, Hernando-Amado S, Reales-Calderon J, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Blanca Sanchez M, Martinez J. 2016. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole K. 2008. Bacterial multidrug efflux pumps serve other functions. Microbe 3:179–185. [Google Scholar]
- 12.Alvarez-Ortega C, Olivares J, Martinez JL. 2013. RND multidrug efflux pumps: what are they good for? Front Microbiol 4:7. doi: 10.3389/fmicb.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, Deng Z, Yan A. 2014. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun 453:254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 14.Piddock L. 2006. Multidrug-resistance efflux pumps: not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 15.Fernando DM, Kumar A. 2013. Resistance-nodulation-division multidrug efflux pumps in Gram-negative bacteria: role in virulence. Antibiotics 2:163–181. doi: 10.3390/antibiotics2010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webber MA, Bailey AM, Blair JMA, Morgan E, Stevens MP, Hinton JCD, Ivens A, Wain J, Piddock LJV. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol 191:4276–4285. doi: 10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckley A, Webber M, Cooles S, Randall L, La Ragione R, Woodward M, Piddock L. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 18.Hirakata Y, Srikumar R, Poole K, Gotoh N, Suematsu T, Kohno S, Kamihira S, Hancock R, Speert D. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J Exp Med 196:109–118. doi: 10.1084/jem.20020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padilla E, Llobet E, Domenech-Sanchez A, Martinez-Martinez L, Antonio Bengoechea J, Alberti S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavri A, Mozina SS. 2012. Involvement of efflux mechanisms in biocide resistance of Campylobacter jejuni and Campylobacter coli. J Med Microbiol 61:800–808. doi: 10.1099/jmm.0.041467-0. [DOI] [PubMed] [Google Scholar]
- 21.Siddiqui R, Khan NA. 2012. Biology and pathogenesis of Acanthamoeba. Parasites Vectors 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandstrom G, Saeed A, Abd H. 2011. Acanthamoeba-bacteria: a model to study host interaction with human pathogens. Curr Drug Targets 12:936–941. doi: 10.2174/138945011795677845. [DOI] [PubMed] [Google Scholar]
- 23.Thomas V, McDonnell G, Denyer SP, Maillard J. 2010. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev 34:231–259. doi: 10.1111/j.1574-6976.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 24.Cirillo J, Cirillo S, Yan L, Bermudez L, Falkow S, Tompkins L. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect Immun 67:4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker J, Brown MRW. 1995. Speculations on the influence of infecting phenotype on virulence and antibiotic susceptibility of Legionella pneumophila. J Antimicrob Chemother 36:7–21. doi: 10.1093/jac/36.1.7. [DOI] [PubMed] [Google Scholar]
- 26.Winiecka-Krusnell J, Linder E. 2001. Bacterial infections of free-living amoebae. Res Microbiol 152:613–619. doi: 10.1016/S0923-2508(01)01240-2. [DOI] [PubMed] [Google Scholar]
- 27.Tosetti N, Croxatto A, Greub G. 2014. Amoebae as a tool to isolate new bacterial species, to discover new virulence factors and to study the host-pathogen interactions. Microb Pathog 77:125–130. doi: 10.1016/j.micpath.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Siddiqui R, Khan NA. 2012. Acanthamoeba is an evolutionary ancestor of macrophages: a myth or reality? Exp Parasitol 130:95–97. doi: 10.1016/j.exppara.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Bare J, Sabbe K, Van Wichelen J, van Gremberghe I, D'hondt S, Houf K. 2009. Diversity and habitat specificity of free-living protozoa in commercial poultry houses. Appl Environ Microbiol 75:1417–1426. doi: 10.1128/AEM.02346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiqui R, Khan NA. 2012. War of the microbial worlds: who is the beneficiary in Acanthamoeba-bacterial interactions? Exp Parasitol 130:311–313. doi: 10.1016/j.exppara.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 31.Vieira A, Seddon AM, Karlyshev AV. 2015. Campylobacter-Acanthamoeba interactions. Microbiology 161:933–947. doi: 10.1099/mic.0.000075. [DOI] [PubMed] [Google Scholar]
- 32.Axelsson-Olsson D, Waldenstrom J, Broman T, Olsen B, Holmberg M. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl Environ Microbiol 71:987–992. doi: 10.1128/AEM.71.2.987-992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snelling W, McKenna J, Lecky D, Dooley J. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl Environ Microbiol 71:5560–5571. doi: 10.1128/AEM.71.9.5560-5571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsson DA, Ellström P, Waldenström J, Olsen B. 2007. Acanthamoeba-Campylobacter co-culture as a novel method for enrichment of Campylobacter species. Zoonoses Public Health 54(Suppl s1):10. [Google Scholar]
- 35.Snelling WJ, Stern NJ, Lowery CJ, Moore JE, Gibbons E, Baker C, Dooley JSG. 2008. Colonization of broilers by Campylobacter jejuni internalized within Acanthamoeba castellanii. Arch Microbiol 189:175–179. doi: 10.1007/s00203-007-0303-0. [DOI] [PubMed] [Google Scholar]
- 36.Axelsson-Olsson D, Olofsson J, Svensson L, Griekspoor P, Waldenstrom J, Ellstrom P, Olsen B. 2010. Amoebae and algae can prolong the survival of Campylobacter species in co-culture. Exp Parasitol 126:59–64. doi: 10.1016/j.exppara.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Axelsson-Olsson D, Svensson L, Olofsson J, Salomon P, Waldenstrom J, Ellstrom P, Olsen B. 2010. Increase in acid tolerance of Campylobacter jejuni through coincubation with amoebae. Appl Environ Microbiol 76:4194–4200. doi: 10.1128/AEM.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olofsson J, Axelsson-Olsson D, Brudin L, Olsen B, Ellström P. 2013. Campylobacter jejuni actively invades the amoeba Acanthamoeba polyphaga and survives within non digestive vacuoles. PLoS One 8:e78873. doi: 10.1371/journal.pone.0078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griekspoor P, Olofsson J, Axelsson-Olsson D, Waldenstrom J, Olsen B. 2013. Multilocus sequence typing and FlaA sequencing reveal the genetic stability of Campylobacter jejuni enrichment during coculture with Acanthamoeba polyphaga. Appl Environ Microbiol 79:2477–2479. doi: 10.1128/AEM.02918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bare J, Sabbe K, Huws S, Vercauteren D, Braeckmans K, van Gremberghe I, Favoreel H, Houf K. 2010. Influence of temperature, oxygen and bacterial strain identity on the association of Campylobacter jejuni with Acanthamoeba castellanii. FEMS Microbiol Ecol 74:371–381. doi: 10.1111/j.1574-6941.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 41.Bui XT, Winding A, Qvortrup K, Wolff A, Bang DD, Creuzenet C. 2012. Survival of Campylobacter jejuni in co-culture with Acanthamoeba castellanii: role of amoeba-mediated depletion of dissolved oxygen. Environ Microbiol 14:2034–2047. doi: 10.1111/j.1462-2920.2011.02655.x. [DOI] [PubMed] [Google Scholar]
- 42.Bui XT, Qvortrup K, Wolff A, Bang DD, Creuzenet C. 2012. Effect of environmental stress factors on the uptake and survival of Campylobacter jejuni in Acanthamoeba castellanii. BMC Microbiol 12:232. doi: 10.1186/1471-2180-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dirks BP, Quinlan JJ. 2014. Development of a modified gentamicin protection assay to investigate the interaction between Campylobacter jejuni and Acanthamoeba castellanii ATCC 30010. Exp Parasitol 140:39–43. doi: 10.1016/j.exppara.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Vaerewijck MJM, Bare J, Lambrecht E, Sabbe K, Houf K. 2014. Interactions of foodborne pathogens with free-living protozoa: potential consequences for food safety. Compr Rev Food Sci Food Saf 13:924–944. doi: 10.1111/1541-4337.12100. [DOI] [Google Scholar]
- 45.Stintzi A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J Bacteriol 185:2009–2016. doi: 10.1128/JB.185.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu Y, Wang Y, Wu J, Lei H. 2010. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells and implications for antibiotic resistance. Infect Immun 78:4157–4165. doi: 10.1128/IAI.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drevets D, Canono B, Leenen P, Campbell P. 1994. Gentamicin kills intracellular Listeria monocytogenes. Infect Immun 62:2222–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsinghorst EA. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol 236:405–420. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 49.Karlyshev A, Wren B. 2001. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using alcian blue dye. J Clin Microbiol 39:279–284. doi: 10.1128/JCM.39.1.279-284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronowski C, James CE, Winstanley C. 2014. Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol Lett 356:8–19. doi: 10.1111/1574-6968.12488. [DOI] [PubMed] [Google Scholar]
- 51.Baugh S, Phillips CR, Ekanayaka AS, Piddock LJV, Webber MA. 2014. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J Antimicrob Chemother 69:673–681. doi: 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- 52.Joshua GWP, Guthrie-Irons C, Karlyshev AV, Wren BW. 2006. Biofilm formation in Campylobacter jejuni. Microbiology 152:387–396. doi: 10.1099/mic.0.28358-0. [DOI] [PubMed] [Google Scholar]
- 53.Lugert R, Gross U, Zautner AE. 2015. Campylobacter jejuni: components for adherence to and invasion of eukaryotic cells. Berl Munch Tierarztl Wochenschr 128:90–97. [PubMed] [Google Scholar]
- 54.Lehri B, Kukreja K, Vieira A, Zaremba M, Bonney K, Karlyshev AV. 2015. Specific genetic features of Campylobacter jejuni strain G1 revealed by genome sequencing. FEMS Microbiol Lett 362:fnu064. doi: 10.1093/femsle/fnu064. [DOI] [PubMed] [Google Scholar]
- 55.Goni P, Fernandez MT, Rubio E. 2014. Identifying endosymbiont bacteria associated with free-living amoebae. Environ Microbiol 16:339–349. doi: 10.1111/1462-2920.12363. [DOI] [PubMed] [Google Scholar]
- 56.Backert S, Hofreuter D. 2013. Molecular methods to investigate adhesion, transmigration, invasion and intracellular survival of the foodborne pathogen Campylobacter jejuni. J Microbiol Methods 95:8–23. doi: 10.1016/j.mimet.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 57.Lambrecht E, Bare J, Chavatte N, Bert W, Sabbe K, Houf K. 2015. Protozoan cysts act as a survival niche and protective shelter for foodborne pathogenic bacteria. Appl Environ Microbiol 81:5604–5612. doi: 10.1128/AEM.01031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anacarso I, de Niederhaeusern S, Messi P, Guerrieri E, Iseppi R, Sabia C, Bondi M. 2012. Acanthamoeba polyphaga, a potential environmental vector for the transmission of food-borne and opportunistic pathogens. J Basic Microbiol 52:261–268. doi: 10.1002/jobm.201100097. [DOI] [PubMed] [Google Scholar]
- 59.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su C, Wang B, Wang S, Wu C, Yu E, Zhang Q, Shen J. 2016. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 7:e01543-16. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, Trust TJ, Guerry P. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun 68:4384–4390. doi: 10.1128/IAI.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, Galan JE. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun 74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karlyshev A, Linton D, Gregson N, Wren B. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473–480. doi: 10.1099/00221287-148-2-473. [DOI] [PubMed] [Google Scholar]
- 63.Gregson N, Rees J, Hughes R. 1997. Reactivity of serum IgG anti-GM1 ganglioside antibodies with the lipopolysaccharide fractions of Campylobacter jejuni isolates from patients with Guillain-Barré syndrome (GBS). J Neuroimmunol 73:28–36. doi: 10.1016/S0165-5728(96)00162-2. [DOI] [PubMed] [Google Scholar]
- 64.Sifre E, Salha BA, Ducournau A, Floch P, Chardon H, Megraud F, Lehours P. 2015. EUCAST recommendations for antimicrobial susceptibility testing applied to the three main Campylobacter species isolated in humans. J Microbiol Methods 119:206–213. doi: 10.1016/j.mimet.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 65.Baldvinsson SB, Sorensen MCH, Vegge CS, Clokie MRJ, Brondsted L. 2014. Campylobacter jejuni motility is required for infection of the flagellotropic bacteriophage F341. Appl Environ Microbiol 80:7096–7106. doi: 10.1128/AEM.02057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.