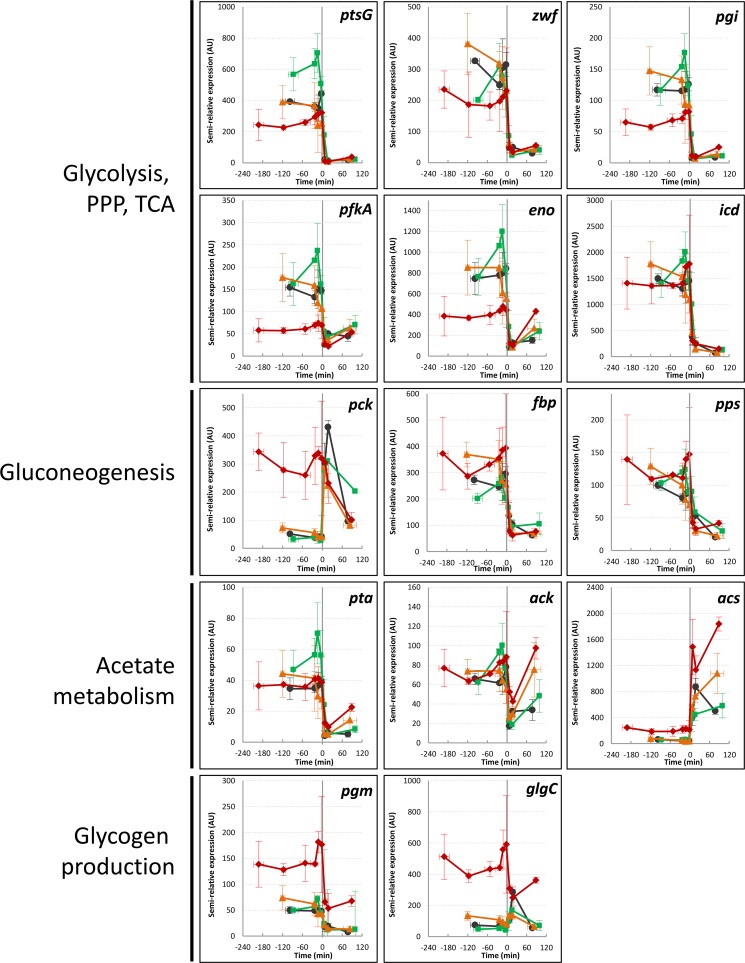
FIG 2 .
Expression of key metabolic genes during the glucose-acetate transition of Csr system mutants. Gene expression (relative to the idnT RNA reference) was investigated from 3 h before to 2 h after glucose exhaustion. WT, black circles; csrBC mutant, green squares; csrD mutant, orange triangles; csrA51 mutant, red diamonds. ptsG, glucose phosphotransferase system permease PtsG subunit (European Bioinformatics Institute European Nucleotide Archive accession no. EG10787); pgm, phosphoglucomutase (accession no. EG12144); glgC, glucose-1-phosphate adenylyltransferase (accession no. EG10379); zwf, glucose 6-phosphate-1-dehydrogenase (accession no. EG11221); pgi, phosphoglucose isomerase (accession no. EG10702); pfkA, phosphofructokinase (accession no. EG10699); eno, enolase (accession no. EG10258); icd, isocitrate dehydrogenase (accession no. EG10489); pck, phosphoenolpyruvate carboxykinase (accession no. EG10688); pta, phosphate acetyltransferase (accession no. EG20173); ack, acetate kinase (accession no. EG10027); ACS, acetyl coenzyme A synthetase (accession no. EG11448); pck, phosphoenolpyruvate carboxykinase (accession no. EG10688); fbp, fructose 1,6-bisphosphatase (accession no. EG10283). The values are averages of three replicates with error bars representing standard deviations. PPP, pentose phosphate pathway; TCA, tricarboxylic acid cycle.