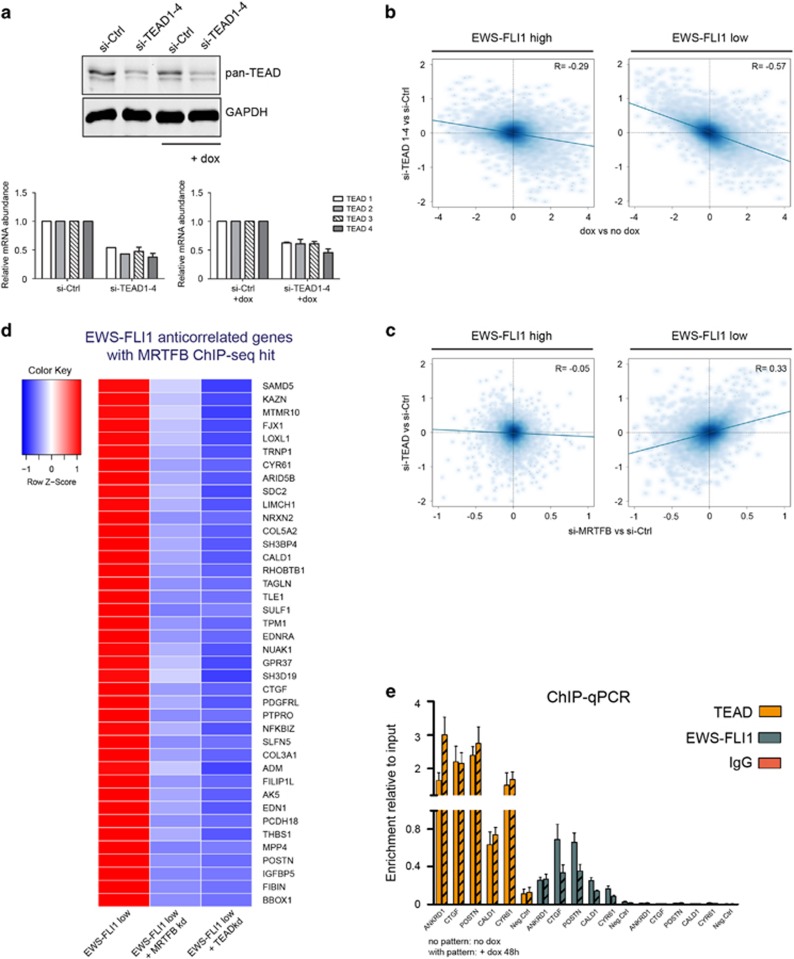
Figure 5.
Effect of TEAD transcription factors on the EWS-FLI1-regulated transcriptome. (a) Western blot analysis showing combined knockdown of all four TEAD transcription factors (TEAD1–4) using siRNA targeting each individual TEAD mRNA. TEAD1–4 factors are recognized by a pan-TEAD antibody. Relative mRNA levels demonstrate that the knockdown efficiency of the four TEAD factors is comparable. Mean±s.e.m. of two biological replicates is shown. (b) Scatter plots showing the effects of the TEAD1–4 knockdown on EWS-FLI1-regulated genes upon EWS-FLI1-high (left plot; R=−0.29) and EWS-FLI1-low (right plot; R=−0.57). TEAD1–4 knockdown antagonizes the effect of EWS-FLI1 depletion, however, more potently upon EWS-FLI1 low conditions. (c) Comparison of gene expression changes induced by MRTFB knockdown (y axis) versus TEAD1–4 depletion (x axis) upon high (left scatter plot; R=−0.05) or low (right scatter plot; R=0.33) EWS-FLI1 levels. Effects of the MRTFB and TEAD1–4 knockdown correlate upon EWS-FLI1-low state only. (d) Heatmap of ‘inversely regulated’ EWS-FLI1/MRTFB target genes (genes for which expression is rescued after the combined EWS-FLI1/MRTFB or EWS-FLI1/TEAD knockdown in comparison to the single EWS-FLI1 knockdown), which were further filtered according to the presence of significant MRTFB ChIP-seq hits, and the presence of TEAD-binding motifs. Gene expression cutoff: EWS-FLI1 knockdown (dox versus no dox) |logFC|⩾1, P<0.05; MRTFB or TEAD knockdown upon EWS-FLI1-low conditions (si-MRTFB+dox/si-TEAD+dox versus si-Ctrl+dox): |logFC|⩽0.4, P<0.1. (e) ChIP–qPCR of for TEAD, FLI1 and IgG ChIP for selected ‘inversely regulated’ genes. ANKRD1 was used as a positive control for the TEAD ChIP–qPCR. Mean±s.e.m. of three biological replicates is shown.