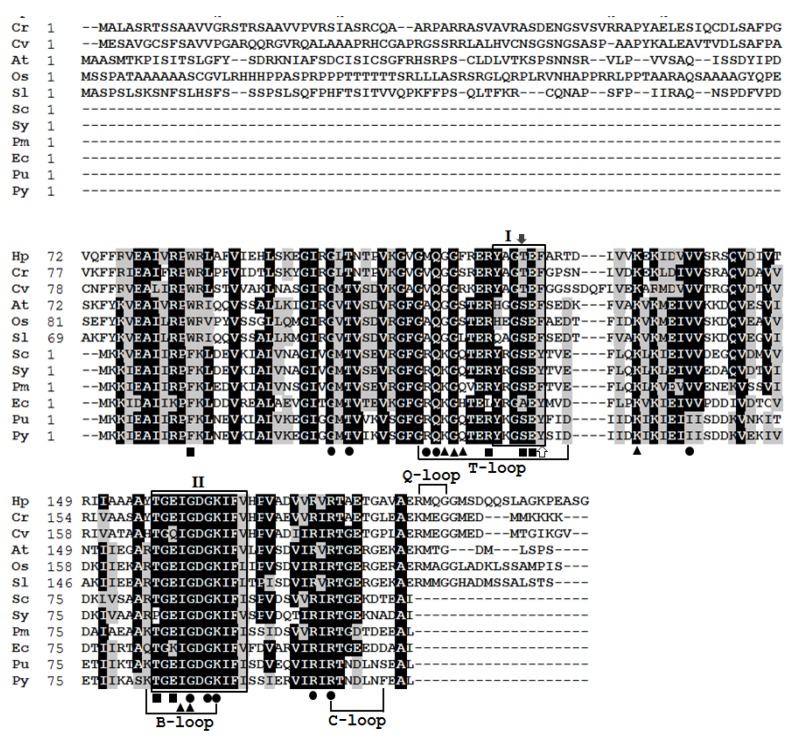
Figure 4.
Alignment of the amino acid sequences of PII proteins among different organisms: Haematococcus pluvialis (Hp; AOO85416), Chlamydomonas reinhardtii (Cr; EDO96407.1), Chlorella variabilis (Cv; AHW46897.1), Arabidopsis thaliana (At; AAC78333.1), Oryza sativa (Os; NP_001054562.1), Solanum lycopersicum (Sl; NP_001234506.1), Synechocystis sp. PCC 6803 (Sc; WP_010873156.1), Synechococcus sp. (Sy; AAA27312.1), Prochlorococcus marinus (Pm; WP_036930683.1), Escherichia coli (Ec; CDZ21367.1), Porphyra umbilicalis (Pu; AFC39923.1), Pyropia yezoensis (Py; YP_536935.1). Residues in black represent >60% identity of aligned PII proteins. Amino acids shaded with grey display similar residues. Box I and box II are PII signature patterns I and II. The white arrow indicates the residue of the corresponding uridylylated threonyl-residue in proteobacteria. The black arrow indicates the residue of corresponding phosphorylated serine-residue in cyanobacteria. Black dots, squares, and triangles show the ATP-, NAGK-, and 2KG-binding residues, respectively.