Figure 1. METTL16 crosslinks to the U6 snRNA in vivo .
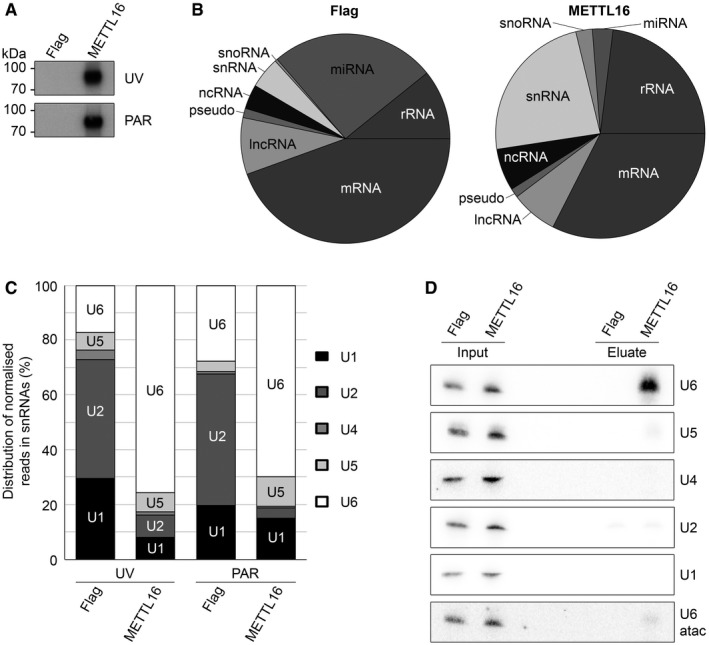
- HEK293 cells expressing METTL16‐Flag (METTL16) or the Flag tag (Flag) were crosslinked using UV light at 254 nm (UV) or grown in the presence of 4‐thiouridine prior to crosslinking at 365 nm (PAR). Protein–RNA complexes were affinity‐purified and bound RNAs trimmed, labelled at the 5′ end with 32P and ligated to sequencing adapters. Protein–RNA complexes were separated by polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and visualised by autoradiography.
- RNAs isolated from the membranes described in (A) were subjected to reverse transcription and PCR amplification to generate cDNA libraries that were analysed by Illumina deep sequencing. The obtained sequence reads were mapped to the human genome and the relative proportion of UV‐CRAC reads mapping to different classes of RNAs is shown.
- The relative distribution of reads mapping to different snRNAs in the UV‐ and PAR‐CRAC data sets derived from cells expressing METTL16‐Flag or the Flag tag is shown. Only snRNAs to which > 5% of the total snRNA hits mapped are labelled within the columns.
- HEK293 cells expressing METTL16‐Flag or the Flag tag were UV crosslinked in vivo and protein–RNA complexes were isolated under denaturing conditions. Crosslinked RNAs were isolated, and inputs (0.2%) and eluates were analysed by Northern blotting using probes hybridising to the snRNAs indicated.
