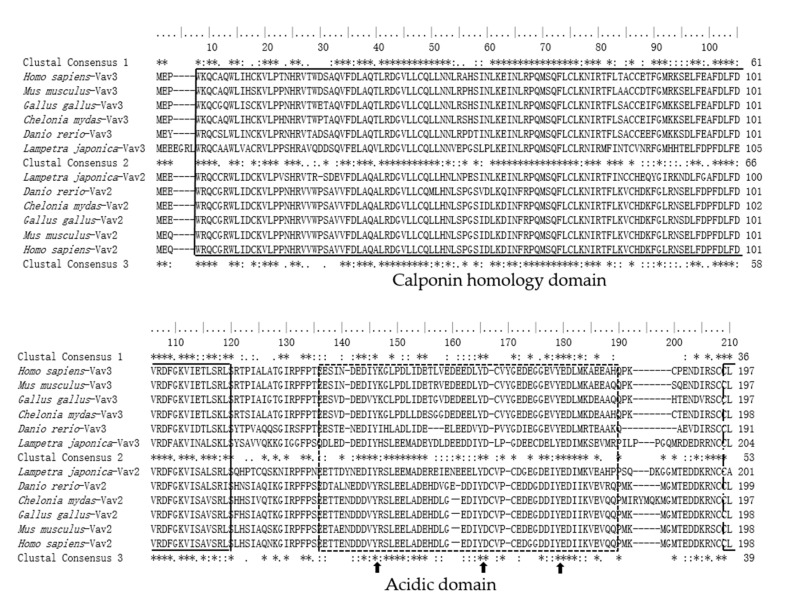
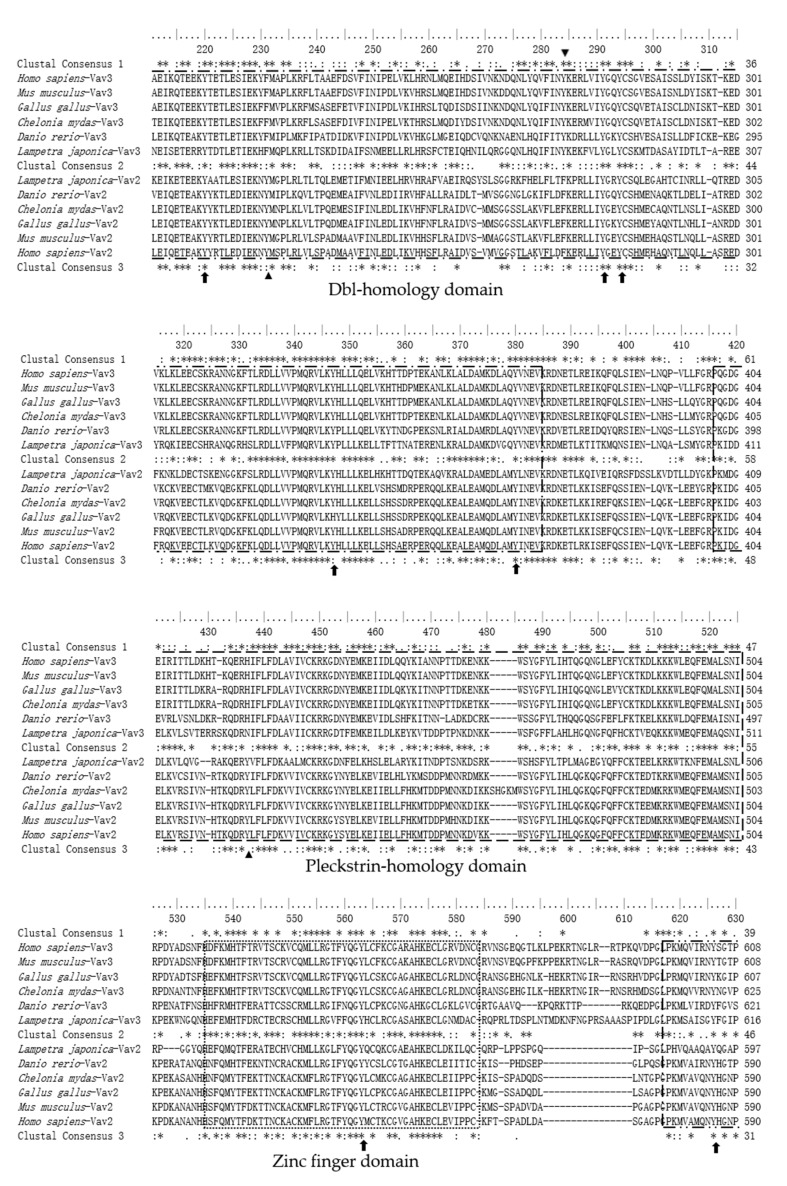
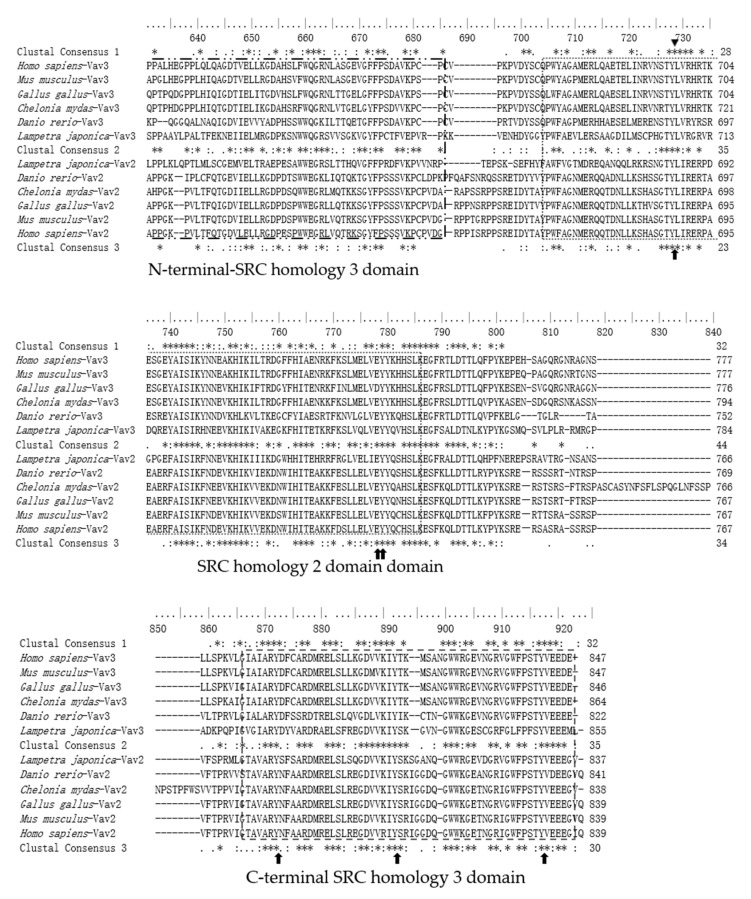
Figure 1.
Multiple sequence alignment analyses of lamprey Vav3 protein sequence with several Vav3 and Vav2 molecules of jawed vertebrates. The accession numbers of these protein sequences are listed below. Homo sapiens-Vav3: AAD20349.1; Mus musculus-Vav3: NP_065251.2; Gallus gallus-Vav3: NP_996745.1; Chelonia mydas-Vav3: XP_007055271; Danio rerio-Vav3: NP_001119865.1; Lampetra japonica-Vav3: KX911208; Lampetra japonica-Vav2: gbAIN44441; Danio rerio-Vav2: XP_009293636.1; Chelonia mydas-Vav2: XP_007061074; Gallus gallus-Vav2: NP_989473.1; Mus musculus-Vav2: XP_006497921.1; and Homo sapiens-Vav2:NP_003362.2. The calponin homology (CH), acidic (Ac), Dbl-homology (DH), pleckstrin homology (PH), zinc finger (ZF), N-terminal SRC homology 3 (NSH3), SRC homology 2 (SH2) and C-terminal SH3 (CSH3) domains are marked with solid line, dashed line, long dash-dot line, long dashed line, dotted line, long dash-dot-dot line, double dashed line and double long dashed line, respectively. The conserved tyrosine residues between Vav3 and Vav2 molecules are indicated by arrows. The conserved tyrosine residues specific to Vav3 or Vav2 are marked by triangles and inverted triangles, respectively. The Clustal Consensus 1, 2 and 3 indicate the identical (*), highly homologous (:) and homologous (.) residues between lamprey Vav3 and Vav3 molecules of jawed vertebrates, between lamprey Vav3 and lamprey Vav2, or between lamprey Vav3 and Vav2 molecules of jawed vertebrates, respectively.