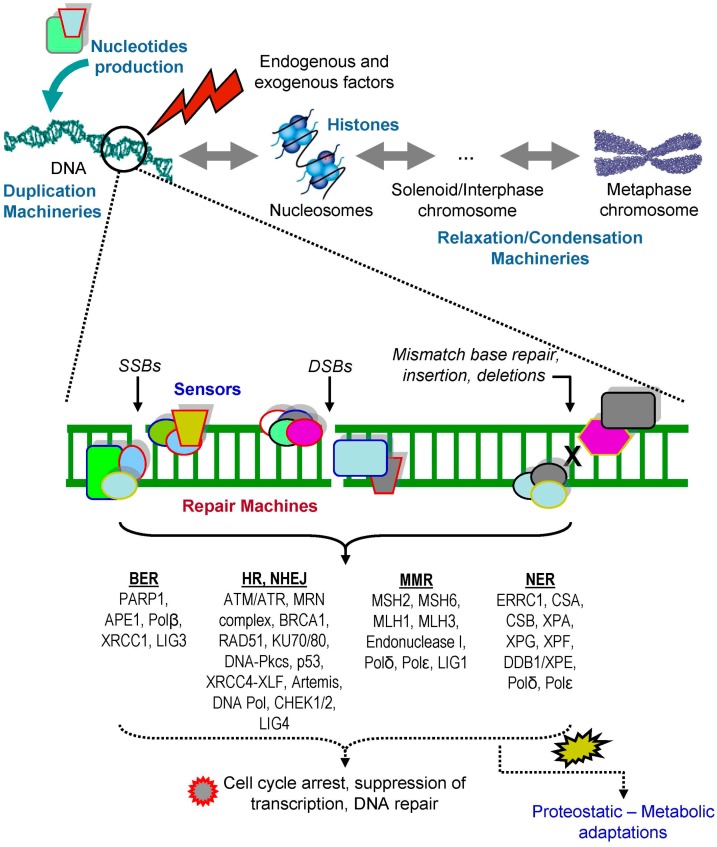
Figure 1.
Protein machines in genome maintenance and repair. Optimum functionality of the proteostasis network impacts on all levels of genome stability as it affects both the physiological processes of nucleotides production, DNA duplication and/or relaxation-condensation, as well as DNA damage sensing and repair. Endogenous and exogenous factors can induce DNA damage in multiple ways such as single strand breaks (SSBs), double strand breaks (DSBs), single or short-patch base lesions and DNA base mismatches. SSBs repair is executed via the base excision repair (BER) pathway (involving, among others, PARP1/2 and XRCC1), while DSBs repair mobilizes the homologous recombination (HR; key players here are the ATM/ATR kinases, the Mre11-Rad50-Nbs1 (MRN) complex, BRCA1/2 and RAD51) and the non-homologous end joining (NHEJ; includes Ku70/80, DNA-PKcs and XRCC4-XLF) pathways. Once the cell has sensed DSBs, the DNA repair machinery is recruited to the lesion in relation to the cell cycle stage; in G1 phase cells undergo repair predominantly through the NHEJ repair pathway, whereas in G2/M the presence of replicated DNA allows the repair through the HR pathway. The mismatch repair pathway (MMR) is executed (among others) via the MSH2/6, MLH1, PMS2 and Exonuclease 1 proteins, while UV-induced DNA lesions are effectively repaired by the nucleotide excision repair (NER). DDR triggers downstream actions (e.g., via CHEK1, CHEK2 and p53 activation) that suppress transcription and cell cycle progression or trigger apoptosis if the damage is not repairable. Likely DDR also induces a number of proteostatic and/or metabolic adaptations, which remain not well understood. It is thus evident that DNA integrity and stability depends heavily on the functionality of its curating protein machines.