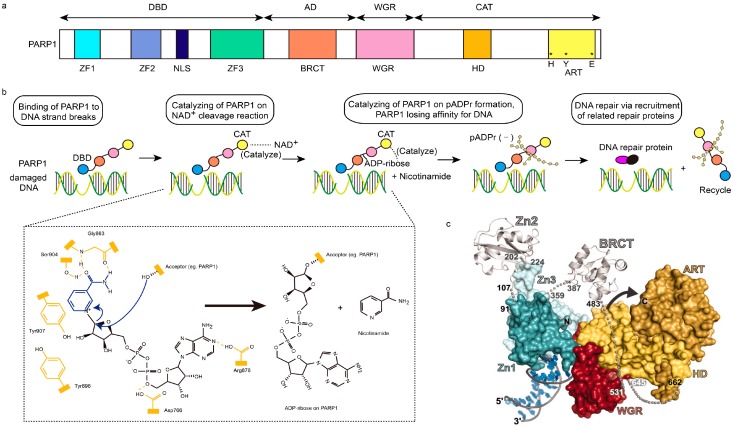
Figure 1.
Structural characteristics of PARP1 and PARP1-based DNA repair. (a) Schematic representation of human PARP1 molecular architecture; (b) Structural process of PARP1-mediated DNA repair. After DNA-binding domain (DBD, blue ball) of PARP1 senses and binds damaged DNA, the NAD+ will be cleaved into ADP-ribose and nicotinamide, which are catalyzed by the catalytic domain (CAT, yellow ball) of PARP1. Then, poly (ADP-ribose) (pADPr) is synthesized on the acceptor PARP1 through the combination of ADP-ribose, assisted by the catalysis of CAT too. Subsequently, PARP1 leaves damaged DNA, due to the dense negative charge of pADPr, allowing the recruitment of related repair proteins and replication. In chemical formulae, domains in yellow represents active domains of PARP1, while blue and black parts represent NAD+. Hydroxyl of Ser904 and carbonyl and NH group of Gly863 in PARP1 interact with the amide moiety of NAD+ via hydrogen bonding interaction, while Tyr907 of PARP1 via π–π stacking interaction. The curved arrows in blue represent the nucleophilic attack by the 2′-hydroxyl of the acceptor site in PARP1, and the release of the nicotinamide procedures; AD, orange ball; WGR, red ball. (c) The PARP1/damaged DNA crystal structure. Zn1, dark green; Zn2, silver (left); Zn3, light green; BRCT, silver (right); ART, dark yellow; HD, light yellow; red, WGR [4,18,19].