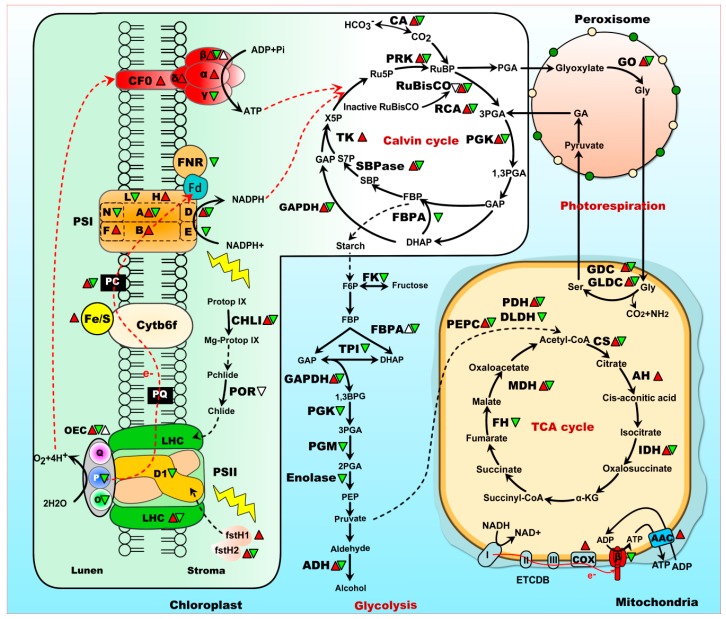
Figure 2.
Schematic representation of heat-responsive photosynthesis and carbon metabolism revealed from proteomics. Red triangle and green triangle represent heat-increased protein abundance and heat-decreased protein abundance, respectively. White regular triangle and white inverted triangles represent heat-increased phosphorylation level and heat-decreased phosphorylation level, respectively. The solid line indicates single-step reactions, and the dashed line indicates multi-step reactions. ADH, alcohol dehydrogenase; AH, aconitate hydratase; AMY, β-amylase; CA, carbonic anhydrase; CHLI, magnesium-chelatase subunit; COX, cytochrome c oxidase assembly protein; Cytb6f, cytochrome b6/f complex; D1, photosystem II reaction-center D1 protein; DLDH, dihydrolipoyl dehydrogenase; FBPA, fructose-bisphosphate aldolase; Fe/S, Rieske Fe/S protein of cytochrome b6/f complex; FK, fructokinase; FNR, ferredoxin—NADP reductase; FPK, fructose-6-phosphate-2-kinase; fstH, filamentation temperature-sensitive H; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GDC, glycine decarboxylase; GLDC, glycine dehydrogenase; GO, glycolate oxidase; IDH, isocitrate dehydrogenase; LHC, chlorophyll a/b binding protein; MDH, malate dehydrogenase; NDUFV, NADH2 dehydrogenase; OEC, oxygen-evolving enhancer protein; PDH, pyruvate dehydrogenase; PEP, phosphoenolpyruvate carboxylase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; PK, pyruvate kinase; PRK, phosphoribulokinase; PSI, photosystem I; PSII, photosystem II; RCA, ribulose-1,5-bisphosphate carboxylase/oxygenase activase; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; SBPase, sedoheptulose-1,7-bisphosphatase; SHMT, serine hydroxymethyltransferase; SUS, sucrose synthase; TPI, triosephosphate isomerase; UPGase, UDP-glucose pyrophosphorylase.