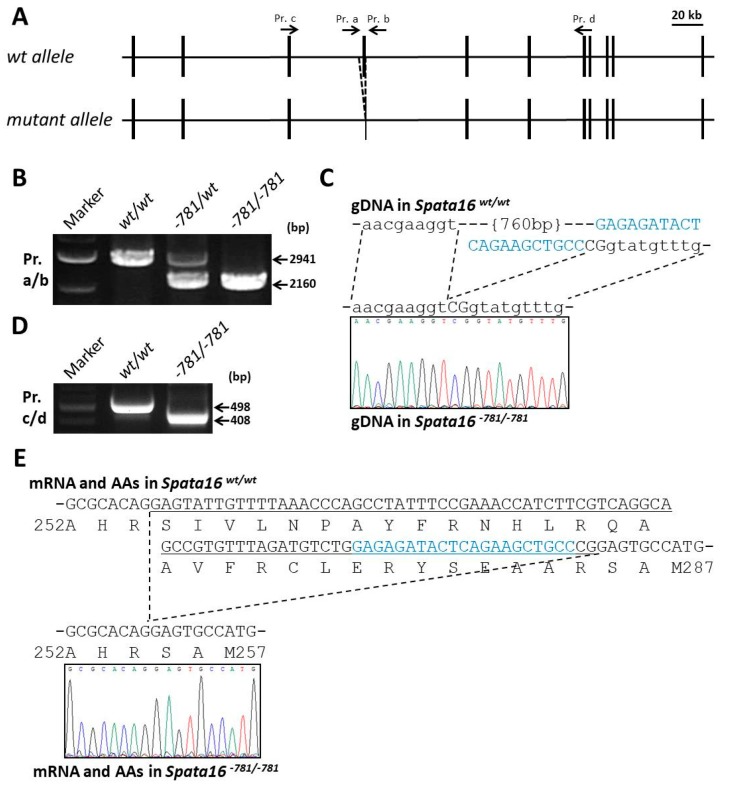
Figure 3.
781-bp deletion of Spata16 by CRISPR/Cas9. (A) Targeting scheme of the 781-bp deletion in the mouse Spata16 locus; (B) The genotyping of Spata16−781/−781 mice by PCR analysis. Both the wild-type allele band at 2941 bp and the deleted allele band at 2160 bp were amplified by PCR; (C) Direct sequencing of the 781-bp deletion around the Spata16 fourth exon. Capital and small letters indicate nucleotides of the exon and intron, respectively. Blue indicates the sgRNA sequence; (D) RT-PCR analysis of a testis in Spata16−781/−781 mice. The 498-bp and 408-bp bands were amplified from wild-type and Spata16−781/−781 mice, respectively; (E) Direct sequencing of Spata16 mRNAs in wild-type and Spata16−781/−781 mice. The 781-bp deletion around the fourth exon in Spata16−781/−781 mice caused the mis-translation of the remaining 2 bp (5′-CG-3′) of the mutant fourth exon. Blue indicates the sequence of the sgRNA.