Fig. 3.
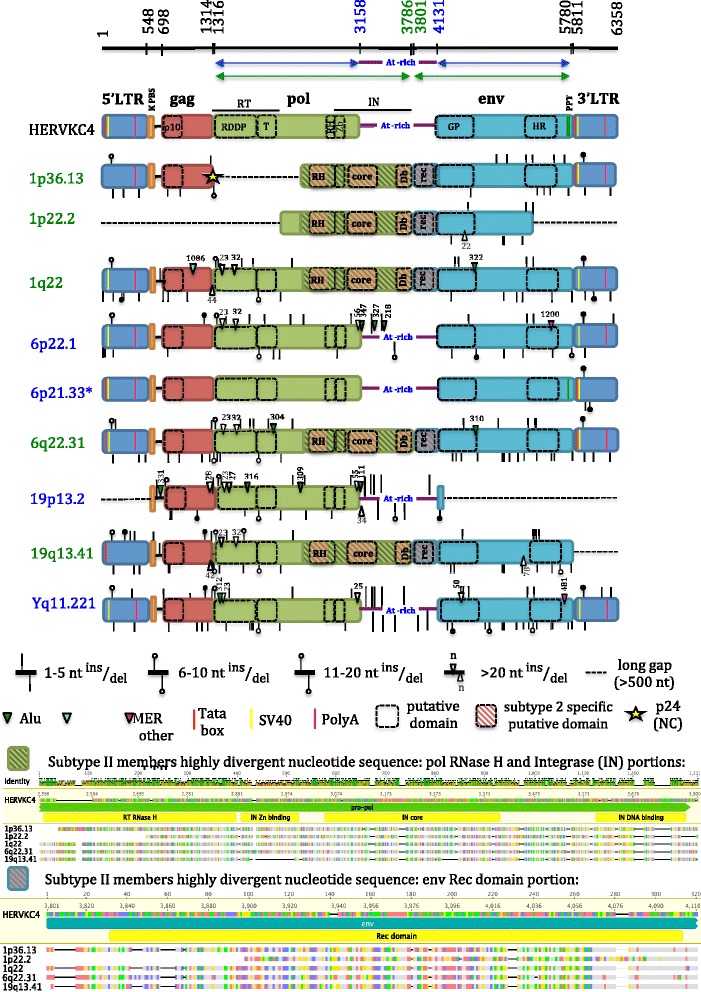
HML10 proviruses structural characterization. Each HML10 provirus nucleotide sequence has been compared to the reference sequence HERV-K(C4) (RepBase). Nucleotides insertions and deletions, LTR regulatory elements and retroviral genes predicted functional domains are annotated. Type II proviruses are reported in red and showed a more divergent nucleotide sequence, especially in pol RNase H and IN portions and env 5′ region (red stripes). Due to the high number of nucleotide changes, the comparison of these portions to the reference is depicted separately. RT: Reverse Transcriptase; RDDP: RNA dependent DNA polymerase; T: thumb; RH: Ribonuclease H; IN: Integrase; Zb: Zinc binding; Db: DNA binding; GP: glycoprotein; HR: Heptad Repeats. Type I proviruses present in the correspondent portion an A/T-rich stretch previously reported for HERV-K(C4) between pol and env genic regions
