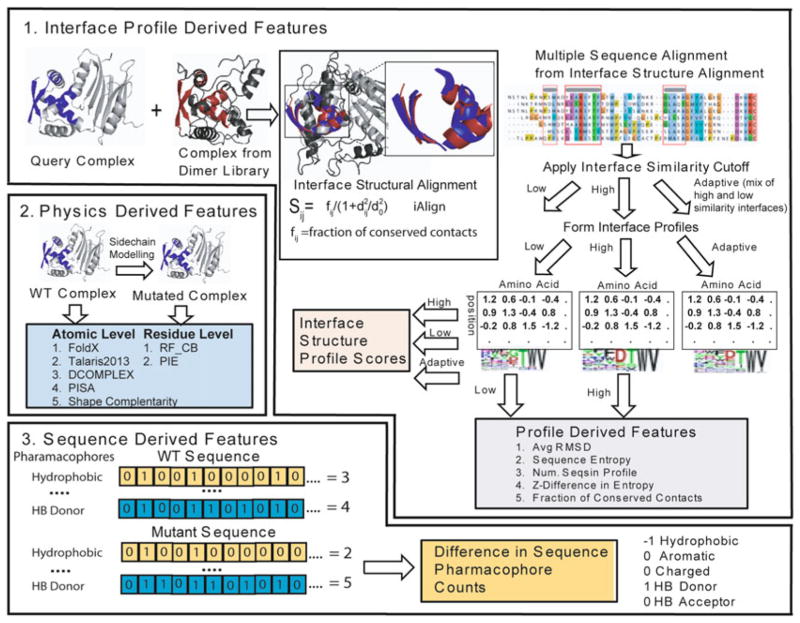
Fig. 2.
Multiscale approach to predicting protein binding affinity using features derived from interface structural profiles, WT and mutant sequences, and physics-based scoring of the structures of the wild-type and mutant complexes. (1) Interface profile scores derived by structural alignment of structurally similar interfaces using an interface similarity cutoff to define the aligned sequences that are used to build the profile. (2) Physics-based scores are formed at the residue or atomic level formed by modeling the mutant monomeric protein and complex and evaluating the difference in energy. (3) Sequence features are formed by the difference between the WT and mutant sequences in the number of hydrophobic (V, I, L, M, F, W, or C), aromatic (Y, F, or W), charged (R, K, D, or E), hydrogen bond acceptors (D, E, N, H, Q, S, T, or Y), and hydrogen bond donating residues (R, K, W, N, Q, H, S, T, or Y) along with difference in amino acid volume calculated from the sequence