Fig. 5. FStitch requires little training data and is robust to low levels of GRO-seq read coverage.
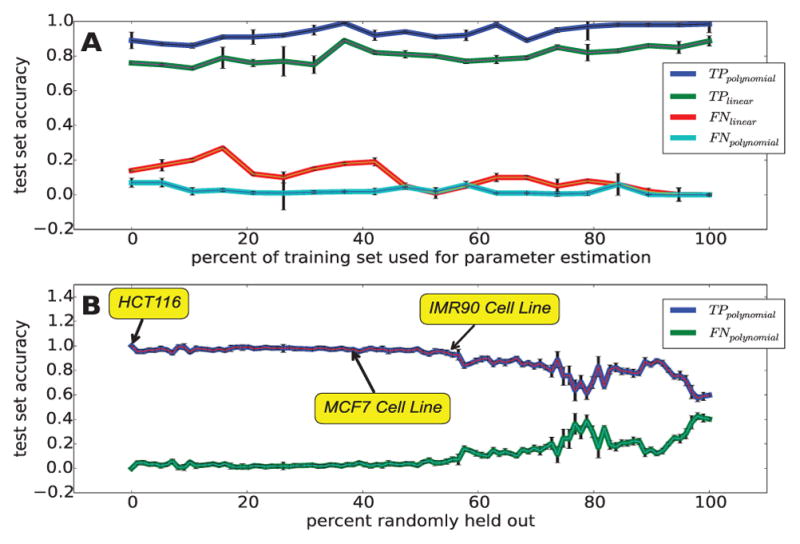
(A) Classification accuracy utilizing successively decreasing amounts of training data to learn feature vector weights, for the polynomial (d = 2 and c = 0; blue and teal) and linear (d = 1 and c = 0; green and red) kernel. (B) Classification accuracy with successively less sequencing depth (dataset size). In this case, we trained on 5% of all available chromosome 1 labels and tested on 50 different subsamples of the curated dataset. T P = true positive rate and F N = false negative rate.
