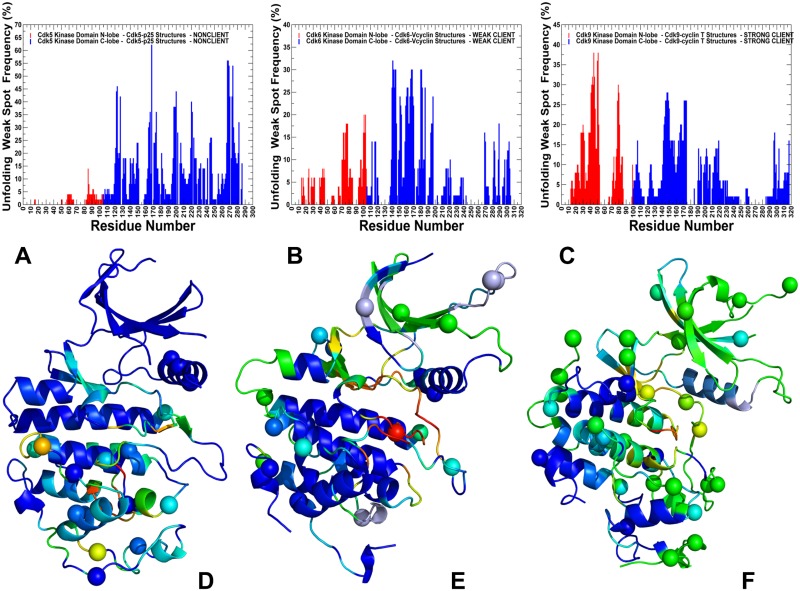
Fig 11. Rigidity analysis of thermal unfolding in the CDK structures.
The frequency of unfolding nuclei (or weak spots) in the CDK5-p25 structure (A), CDK6-Vcyclin (B) and CDK9-cyclin T structure (C). The frequencies of the kinase domain residues are shown in colored bars, with the N-lobe residues in red bars and C-lobe in blue bars. Unfolding nuclei or weak spots are defined as residues that belong to the giant rigid cluster until the folding/unfolding transition point and break away from the giant rigid cluster immediately after transition during the network-based emulation of thermal unfolding. The localization of high frequency weak spot residues characterizes protein stability and rigidity/flexibility partition in the protein structure. The higher the frequency of a weak spot, the more probable unfolding begins from these residues. Structural mapping of predicted weak spots on structures of CDK5 kinase domain from CDK5-p25 complex (D), CDK6 kinase domain from CDK6-Vcyclin complex (E) and CDK9 domain from CDK9-cyclin T complex (F). Crystallographic conformations are colored using a color range from red (highest ranking weak spot) to blue (lowest ranking weak spot). The positions of pathogenic mutations in CDK structures are shown in spheres colored according to weak spot ranking.