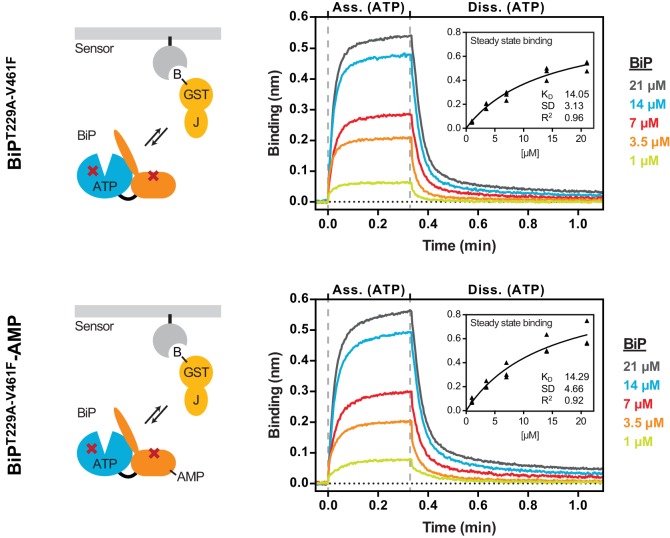
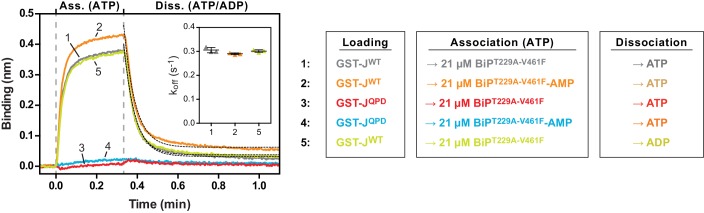
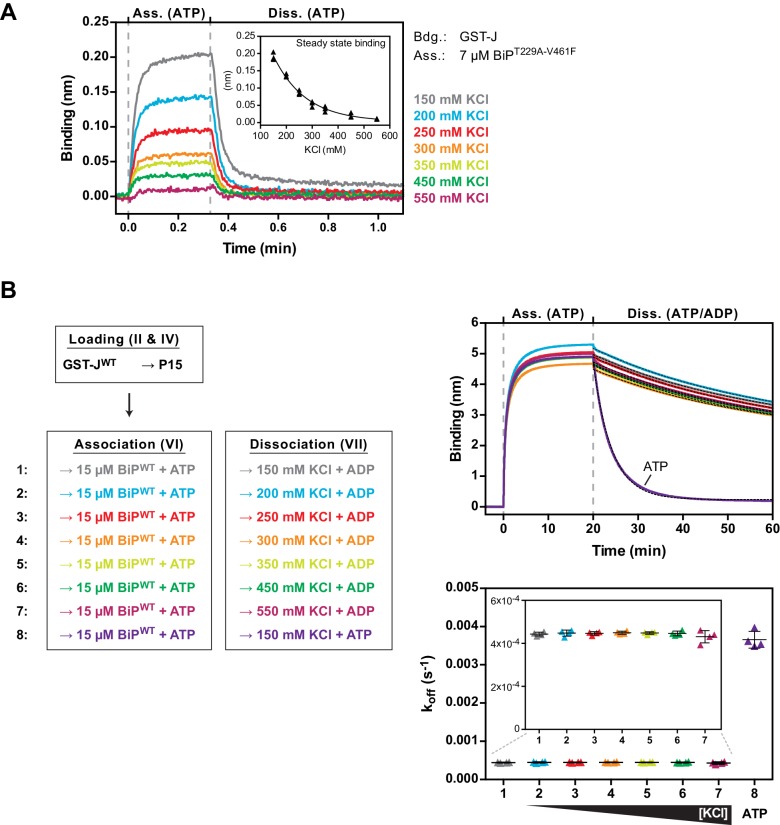
Figure 5. AMPylation does not alter non-substrate interactions between BiP and a model J-domain.
Bio-layer interferometry (BLI) experiment based on the experiment outlined in Figure 4C to measure the functionally relevant non-substrate interactions between an ATPase and substrate binding BiP mutant protein (BiPT229A-V461F) and a model J-domain fused to GST (GST-J). The cartoons on the left depict the anticipated interactions between BiP in solution and the J-domain on the sensor surface during the association step and plots of BLI binding signals against time are shown on the right. After an initial equilibration step the sensors were saturated with biotinylated GST-J followed by another wash step to achieve a stable baseline signal (not shown). The sensors were then exposed to solutions containing different concentrations of unmodified or AMPylated BiPT229A-V461Fproteins to measure their association with the sensors. Dissociation was detected in protein-free solutions containing ATP. Representative plots of recorded binding signals of the association and dissociation steps are shown. The insets present plots of the plateau binding amplitudes during the association step against BiP concentrations of three independent experiments as well as the obtained dissociation constant (KD) values with the corresponding standard deviations and the R2 values of the fits.