Abstract
ADAM12, (A Disintegrin and metalloproteinase domain-containing protein 12), is upregulated in epithelial cancers and contributes to increased tumor proliferation, metastasis and endocrine resistance. However, its role in tumor angiogenesis is unknown. Here we report that ADAM12 is upregulated in the vessels of aggressive breast tumors and exerts key regulatory functions. ADAM12 significantly increases bFGF-mediated angiogenesis in vivo and ADAM12 levels are upregulated in tumors that have undergone a switch to the angiogenic phenotype. Importantly, ADAM12-overexpressing breast tumors display a higher microvessel density (MVD). Our aim was to identify the mechanisms by which tumor-associated ADAM12 promotes angiogenesis. ADAM12 expression in breast tumor cells correlated with a significant upregulation of proangiogenic factors such as VEGF and MMP-9 and downregulation of antiangiogenic factors such as Thrombospondin-1 (THBS1/TSP1) and Tissue inhibitor of metalloproteinases-2 (TIMP-2). Co-culture with ADAM12-expressing tumor cells promoted endothelial cell (EC) recruitment and capillary tube formation. Conversely, downregulation of endogenous ADAM12 in breast cancer cell lines resulted in reduction of pro-angiogenic factors and EC recruitment. These ADAM12-mediated effects are driven by the activation of EGFR, STAT3 and Akt signaling. Blockade of EGFR/STAT3 or silencing of ADAM12 reversed the proangiogenic tumor phenotype, significantly downregulated pro-angiogenic mitogens and reduced EC recruitment. In human breast cancer tissues, ADAM12 expression was significantly positively correlated with pro-angiogenic factors including VEGF and MMP-9 but negatively associated with TSP1.
Keywords: ADAM12, tumor angiogenesis, STAT3
Introduction
Tumor neovascularization is now widely recognized as one of the driving forces for tumor progression, invasion and metastasis. An understanding of the mechanisms that promote tumor angiogenesis could help us target tumors more efficiently and effectively. ADAM12 (a disintegrin and metalloprotease) is upregulated in breast (1–5), prostate (6), ovarian (7), skin (8), stomach (9), lung (10) and brain cancers (11), however, a role for ADAM12 in either developmental or pathological angiogenesis has not yet been described. Human ADAM12 can be expressed as two alternately spliced variants: the membrane-associated ADAM12-L, and the shorter secreted ADAM12-S (12). Both isoforms are composed of pro-, metalloprotease, disintegrin and cysteine-rich domains respectively, and a non-homologous cytoplasmic tail. ADAM12-L has a transmembrane region. ADAM12 contributes to tumor progression and metastasis in both orthotopic and transgenic tumor models by promoting tumor cell proliferation, migration and invasion (3,13,14), inducing tumor cell resistance to apoptosis (4), promoting endocrine resistance (15) and enhancing malignant tumor-stroma crosstalk (6). We and others have shown that circulating ADAM12 may be a prognostic marker for a variety of cancers including breast and prostate cancers (1,16–21), and may predict breast cancer risk (22).
ADAM12 knockout mice are viable and fertile (23), suggesting that it may not play a critical role during developmental angiogenesis, however, in the absence of ADAM12, prostate tumors were significantly smaller in size in the W10 mouse prostate tumor model (6). Similarly, loss of ADAM12 in tumor cells significantly reduces breast tumor progression in the PyMT model (2). Importantly, while ADAM12’s role in tumor cell proliferation and reduced apoptosis has been reported, it remains unknown whether the resulting reduction in tumor growth in ADAM12KO animals was a consequence of reduced tumor angiogenesis. ADAM12 is upregulated in the ovarian and breast tumor vasculature (24–26). In the current study, we show that ADAM12 is elevated in breast tumor vessels compared to normal vessels and that while quiescent endothelium has low or no ADAM12, activated endothelium has significantly upregulated expression. We further demonstrate that tumor cell-associated ADAM12 facilitates tumor neovascularization via dysregulated expression of pro- and anti-angiogenic factors in the tumor microenvironment through the activation of EGFR/STAT3/Akt-dependent pathways. To our knowledge, this is the first report of an ADAM protease contributing to tumor angiogenesis.
Materials and Methods
Reagents
Antibodies used include: ADAM12 (Proteintech Group, Chicago, IL), CD31 (BD Biosciences, San Jose, CA) thrombospondin-1 (Neomarkers, Freemont, CA), phosphoSTAT3/total STAT3, phosphoMAPK/total MAPK, phosphoAkt/totalAkt and phosphoVEGR2/total VEGFR2 (Cell Signaling, Danvers, MA), GAPDH (Millipore, Temecula, CA), HRP-conjugated anti-rabbit and anti-mouse antibodies (Vector Biolabs, Burlingame, CA). Small molecule inhibitors include: EGFR inhibitors AG1478 (CAS #175178-82-2, EMD Millipore, Billerica, MA) and PD15035 (CAS# 183322-45-4, R & D Systems, Minneapolis, MN) STAT3 inhibitors; Stattic (CAS# 19983-44-9, R & D Systems) and Niclosamide (CAS# 50-65-7, Sigma Aldrich, St. Louis, MO). VEGF antibody (Ranibizumab, Genetech, San Francisco, CA).
Cell lines
MCF-7, MDA-MB-231, T47-D and MILE cells were obtained from American type culture collection (ATCC, Manassas, VA) and cultured as per ATCC protocols. Human dermal microvascular endothelial cells (HMVEC) and human umbilical vein endothelial cells (HUVEC) were obtained from Lonza (Walkersville, MD) and cultured as per protocol. Our laboratory tests cultured cells for Mycoplasma contamination every three months. Cells testing positive are discarded or treated with an antibiotic regimen until Mycoplasma-free. The passage number for cells did not exceed P8 after thawing. For EC/tumor cell co-culture studies, 3 x 105 HMVEC were seeded in 6-well plates in EGM-2 medium, next day ADAM12-expressing clones or WT MCF-7 cells were seeded onto cell culture inserts (1 μm; Corning Inc. New York, NY) and placed on top. EC/TC were co-cultured for 24 h in serum-free EBM-2, subsequently, EC lysates were prepared for analysis.
Transfection of cells
HUVEC cells were transiently transfected with pcDNA3 plasmid encoding human full-length ADAM12-L and ADAM12-S using Amaxa Nucleofector Kit (Lonza, Walkersville, MD). For the selection of stable clones (MCF-7, T47-D, MDA-MB-231) clones were selected based on neomycin resistant growth (G418, 0.5 mg/ml; Life Technologies, CA).
Gene knockdown
ADAM12 shRNA lentiviral particles (sc-41414-V, Santa Cruz Biotechnology, Dallas. TX) and control shRNA (sc-108080) were used to engineer stable ADAM12 knockdown in MDA-MB-436 cells according to the manufacturer’s protocol. In brief, tumor cells were seeded at 70% confluence and treated with Polybrene (5 μg/ml) followed by addition of lentiviral particles for 48 h. Stable cells expressing ADAM12 shRNA were selected by puromycin (2.5 μg/ml) for 2–3 weeks. Downregulation of ADAM12 expression in breast tumor cells (BTC) was confirmed via Realtime RT-PCR, immunoblot and ELISA. siGENOME human EGFR siRNA (1956) and human STAT3 siRNA (6774) (Dharmacon, GE Healthcare, Pittsburgh, PA) was used to silence EGFR and STAT3 expression respectively, in breast tumor cells according to manufacturer’s protocol. Downregulation of protein expression was confirmed via immunoblot.
Immunoblotting and ELISA
Cell lysates were prepared using 1X lysis buffer (Cell Signaling Technology, Danvers, MA) and supplemented with phosphatase inhibitor cocktail (phosSTOP, Roche Life sciences, Indianapolis, IN). For immunoblot analysis serum-free conditioned medium was concentrated using 10 kDa cutoff filter (YM-3 Microcon, Millipore) to load 35 μg total protein/lane. Immunoblotting was conducted as described before (1,3). For ELISA neat serum-free conditioned media or 4-fold diluted lysates were used. ELISAs used in the study include: ADAM12, thrombospondin-1, VEGF, MMP-9, TIMP-2 (R & D Systems, Minneapolis, MN). Protein concentration of the lysates and conditioned medium was determined using the Bradford method (Biorad laboratories, Hercules, CA).
Migration and EC recruitment assay
In vitro EC migration studies were conducted using a modified boyden chamber assay as described previously (3). Briefly, EC (104) were plated in the transwell chamber (Costar Transwell Assay; Corning Inc., Corning NY) in EGM-2 medium supplemented with 0.2% FBS. 600 μl of EBM-2 medium was placed in the lower chamber. After 24 h, cells were fixed and stained with Diff-Quik Stain Set (Siemens Inc., Deerfield, IL). Cells adhering to the top of the filter were gently wiped away and those adhering to the lower surface of the filter were counted using a Nikon microscope at 400X magnification (5 fields/filter; n=3 filters per treatment). For endothelial cell (EC) recruitment assays, 2.5 x 104 tumor cells were seeded into 24-well plates 24 h before the start of the assay. HMVEC or HUVEC were serum-starved in EGM-2 media supplemented with 0.2% FBS for 24 h. Tumor cells were washed with PBS and EGM-2 containing 0.2% FBS was added to each well. 5 x 104 EC were plated in the upper transwell chamber as described above. After 18 h, migrated EC were stained and counted as described. For STAT3 and EGFR inhibition tumor cells were treated with Stattic (5 μM) or AG1478 (10 μM) or PD15035 (1 μM) in serum-containing medium overnight. Subsequently, tumor cells were washed with PBS, trypsinized and plated in 24 well plates and EC recruitment was conducted as described above. For combined ADAM12 downregulation and VEGF targeting, MDA-MB-436 control shRNA cells or ADAM12-silenced cells were treated with control mouse IgG (1μg/ml) or VEGF antibody (0.5 μg/ml or 1 μg/ml) and EC recruitment was conducted as described.
Endothelial tube formation assay
Reduced growth factor matrigel (100 μl) was first added to 96-well plate on ice and allowed to polymerize at 37 °C for 1 h. HUVECs (2 x 104; passage 3–6) transiently transfected with control vector, ADAM12-L or ADAM12-S were seeded onto the matrigel in low serum-containing medium (EBM2 + 0.1% FBS) and placed in the incubator for 12 h. Images of tubes were captured using an inverted microscope and the number of cords from four representative wells for each cell type was used for comparison. To analyze the effect of exogenous ADAM12 in this assay indicated concentrations of recombinant human ADAM12-S (furin-activated; R & D Systems) was added to the wells.
Substrate gel electrophoresis
MMP-2 and MMP-9 activity was detected using gelatin zymography as described previously (1). Briefly, serum-free conditioned-medium (40μl) was mixed with sample buffer and resolved via electrophoresis. Substrate digestion was conducted as previously described (3). Gels were stained with Coomassie and imaged using Biorad Imager. Bands of MMP enzyme activity were detected as zones of clearance on a background of uniform blue staining.
Real-time RT-PCR analysis and angiogenesis PCR array
Total RNA was extracted from cells using the RNAeasy Qiagen Kit according to the manufacturer’s protocol (Qiagen, Germantown, MD). cDNA was prepared by reverse transcription from 1μg of total RNA using the Superscript III Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA). Forward and reverse primers are indicated in Supplementary Table I. Real time RT-PCR was performed using iQ™ SYBR™ Supermix (Biorad laboratories). GAPDH was used for normalization. Angiogenesis PCR array was performed to compare the angiogenic profile of ADAM12-expressing MCF-7 cells versus WT MCF-7 using the commercially available RT2 Profiler PCR array for human angiogenesis (PAHS-024; Qiagen, Valencia, CA) and analyzed as described by the manufacturer. Expression of genes that were differentially expressed >2-fold-change were validated via Real time RT-PCR.
Mouse corneal pocket angiogenesis assay
Animal studies were conducted in compliance with the Boston Children’s Hospital IACUC guidelines. For the mouse corneal pocket assay hydron pellets containing sucrose octasulfate and either a low dose of human bFGF (30 ng) plus human recombinant ADAM12-S (80 ng; 4416-AD, R&D Systems; latent ADAM12-S was activated using furin according to the manufacturer’s protocol) or bFGF (30 ng) alone were implanted into corneal micropockets of 8-wk-old male C57Bl/6 mice (Jackson Laboratory, Bar Harbor, ME, USA). On day 7, angiogenesis in each eye was evaluated using a slit-lamp microscope and photographed. The area of neovascularization for each cornea was calculated from the length of the vessels (VL) invading the cornea as well as the clock hours (CH) covered with the formula VL × CH × 0.0628.
Orthotopic breast tumor xenografts
WT MCF-7 and ADAM12-expressing stable clones were cultured until confluent. 4 x 106 cells of each type were suspended in 40 μl cold HBSS and injected into the exposed fourth right inguinal mammary fat pad of 8–10 week old female BALB/c nude mice (Charles River Labs, Wilmington, MA) as previously described (3). Slow-release 17-β-estradiol pellets (Innovative Research of America, Sarasota, FL) were implanted at the time of injection. Tumor volume was calculated based on the formula (lengthxwidthxwidth)/2. Animals were sacrificed when tumor sizes reached ~1 cm in diameter or when mice were moribund. Primary tumors were collected for preparation of frozen sections for analysis. All animal studies were conducted in compliance with the Boston Children’s Hospital IACUC guidelines.
Correlation analysis
ADAM12 expression data for a cohort of human breast tumors (351 human breast tumors: GSE2109; 104 breast tumors: GSE42568 retrieved from GEO) profiled using the Affymetrix® Gene Profiling Array cGMP U133 P2 were analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). The expression values were log2-transformed and median-centered for all tumors, Pearson r and P values are shown for each comparison.
Statistical analysis
Results for migration, tube-formaion and EC recruitment assays are reported as mean ± SD of at least 3 independent experiments. Differences between experimental groups were statistically analyzed using the one way ANOVA test and values of P≤ 0.05 were considered to be statistically significant.
Extended Methods have been provided in Supplemental Data Files.
Results
ADAM12 expression is upregulated in the tumor endothelium
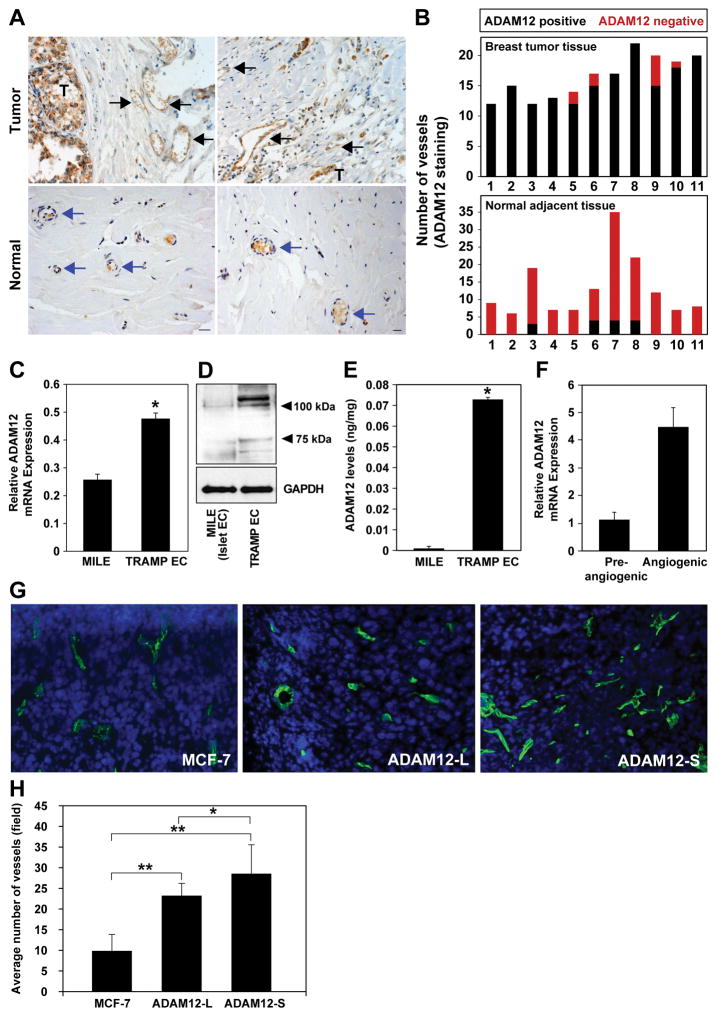
Immunohistochemical (IHC) analyses of ADAM12 in human breast tumors show low or no ADAM12 in normal breast tissue, however significantly higher levels of ADAM12 protein were detected in breast tumor epithelium (Fig. 1A). The endothelium in the vicinity of or within the breast tumors was also positive for ADAM12 (Fig.1A). In contrast, vessels in normal breast tissue did not express ADAM12 (Fig.1A,B). Consistent with these findings, tumor endothelial cells (EC) isolated from TRAMP prostate tumors (27) had significantly higher mADAM12 (mouse ADAM12) mRNA and protein compared to normal mouse islet endothelial cells (MILE; Fig.1C–E). Our current findings are consistent with reports of upregulated ADAM12 in ovarian, breast and skin cancer endothelium (8,24–26). Analysis of ADAM12 gene expression during the switch to the angiogenic phenotype indicated a ~4-fold increase in ADAM12 transcripts in angiogenic versus pre-angiogenic tumors (Fig. 1F) suggesting that ADAM12 may play a role in the acquisition/development of the tumor vasculature in vivo. In addition, the impact of tumor-associated ADAM12 was examined on the tumor vasculature using MCF-7 xenograft tumors. We have previously reported that ADAM12 overexpression in breast tumors results in increased tumor take, tumor size, and metastasis (3). Microvessel density (MVD) of xenograft wild type MCF-7 tumors or those overexpressing ADAM12-S or ADAM12-L isoforms, was analyzed via CD31 staining (Fig. 1G, H). ADAM12-L-expressing tumors displayed a moderate (2.3-fold; P<0.0001) increase in the number of microvessels whereas ADAM12-S-expressing tumors had a significantly higher MVD as compared to MCF-7 tumors (2.9-fold; P<0.0001, Fig 1H). Our results indicate that ADAM12 is increased in tumor-associated endothelium and further suggest that ADAM12 expression in tumor cells may promote angiogenesis in a paracrine manner.
Figure 1. ADAM12 expression is upregulated in tumor endothelium.
Tumor-associated vessels stained positive for ADAM12 (black arrows, T; tumor) compared to vessels in adjacent normal breast tissue which were negative (blue arrows;A). A majority of vessels within or near breast tumors were ADAM12-positive (black bars) compared to normal breast vessels which were largely ADAM12-negative (red bars) (n=11;B). ADAM12 transcript (C) and protein (D; immunoblot, E; ELISA) were significantly higher in EC isolated from prostate tumors (TRAMP model) compared to normal mouse islet EC. GAPDH is included as the loading control (D). Realtime RT-PCR and ELISA results are an average of 3 and 2 independent experiments respectively. ADAM12 mRNA (relative to GAPDH) increased ~4-fold during the switch to the angiogenic phenotype (F). Microvessel density (CD31 staining) in MCF-7, ADAM12-L-expressing and ADAM12-S-expressing breast tumors (n=3 tumors, 6 sections/tumor) (G,H) (one way ANOVA; *P<0.05, **P<0.00001).
ADAM12 expression is upregulated in the activated endothelium
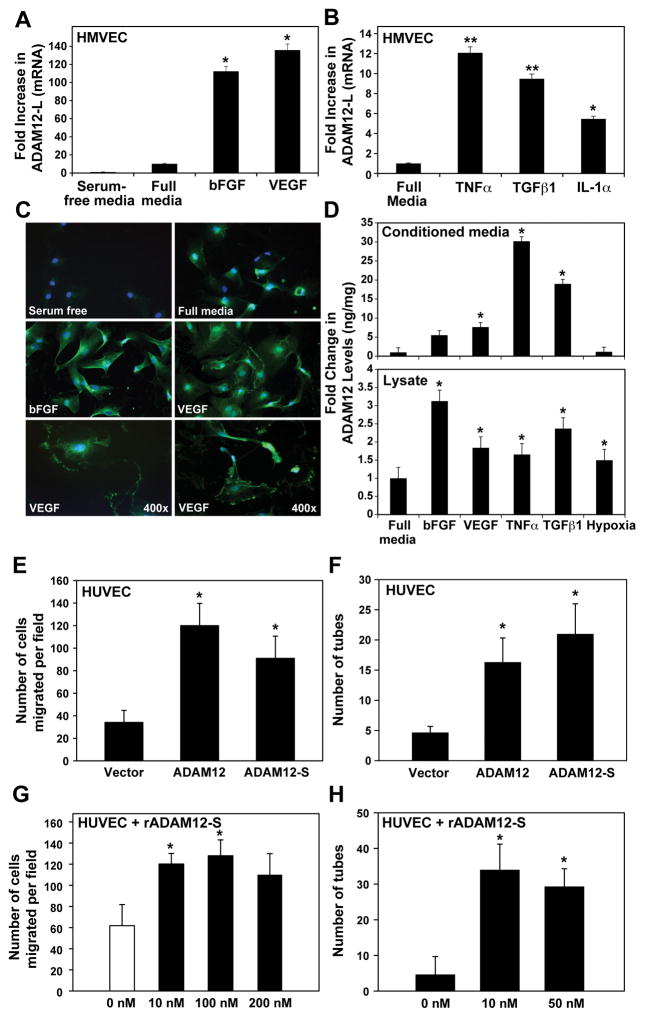
Constitutive expression of ADAM12 was very low in HUVEC (Supplemental Fig. 1) and human dermal microvascular endothelial cells (HMVEC) in vitro (Fig. 2A). Interestingly, ADAM12-L mRNA was upregulated in EC in response to treatment with angiogenic factors such as VEGF and bFGF as well as inflammatory cytokines such as TNFα, TGFβ1 and IL1α (Fig. 2A,B). ADAM12-S mRNA levels are also upregulated in HMVEC by VEGF, TGFβ and TNFα treatment (Suppl. Fig 1A). In addition, immunostaining of HMVECs indicated upregulated ADAM12 levels following bFGF and VEGF treatment (Fig. 2C). In response to VEGF treatment, ADAM12 appeared to strongly localize to the ruffled edges or invadopodia of EC (Fig. 2C, bottom). Significantly higher levels of ADAM12 protein were detected in conditioned medium (CM; ADAM12-S) and lysates (both ADAM12 isoforms) from HMVECs treated with angiogenic factors (VEGF, bFGF) or cytokines (TNFα, TGFβ1) (Fig. 2D). Under hypoxic conditions, there was a slight increase in ADAM12 in cell lysates, however, levels in the CM stayed unchanged (Fig. 2D). These data indicate that while quiescent EC do not express ADAM12, activation of the endothelium via angiogenic and/or inflammatory mediators results in increased expression of ADAM12 in EC.
Figure 2. ADAM12 expression is upregulated in activated endothelial cells and stimulates cell migration and tube-formation.
ADAM12-L transcript (A, B) and protein (immunostaining; C) was higher in HMVECs in response to treatment with bFGF (50ng/ml) and VEGF (100ng/ml) or cytokines like TNFα (5ng/ml), TGFβ-1 (5ng/ml) or IL-1α (1ng/ml). Treatment with angiogenic factors or cytokines resulted in 5–10-fold increase in ADAM12 in the CM (D; top), and lysates (D; bottom). Realtime RT-PCR and ELISA results are an average of 3 and 2 independent experiments respectively. Migration of ADAM12-L and ADAM12-S-expressing HUVECs was significantly higher than vector-transfected cells (E). Tube-formation of EC was significantly increased in ADAM12-expressing cells compared to controls (F). The effect of exogenous recombinant ADAM12-S on migration and tube-formation in EC (G,H) (average of 3 independent experiments, one way ANOVA; *P<0.05, **P<0.00001).
ADAM12 induces migration and tube formation in endothelial cells
To investigate the role of ADAM12 in angiogenesis, we transiently expressed the two isoforms of ADAM12 (-L and –S) in ECs. ADAM12 expression was confirmed by RT-PCR and immunoblot (data not shown). Migration rates of ADAM12-L and ADAM12-S-expressing HUVECs were significantly higher than control vector-transfected cells (Fig. 2E; P<0.0001). Similarly, tube-formation efficiency of HUVEC’s was significantly increased in ADAM12-expressing cells as compared to controls (Fig. 2F; P<0.05). Exogenous addition of recombinant ADAM12-S resulted in significantly increased cell migration (Fig. 2G; P<0.0002) and tube-formation (Fig. 2H; P<0.05). EC proliferation was not impacted by either overexpression or exogenous addition of ADAM12-S (data not shown). Our findings indicate that ADAM12 overexpression or exogenous addition can significantly promote critical EC functions such as migration and tube-formation.
ADAM12 induces angiogenesis in vivo
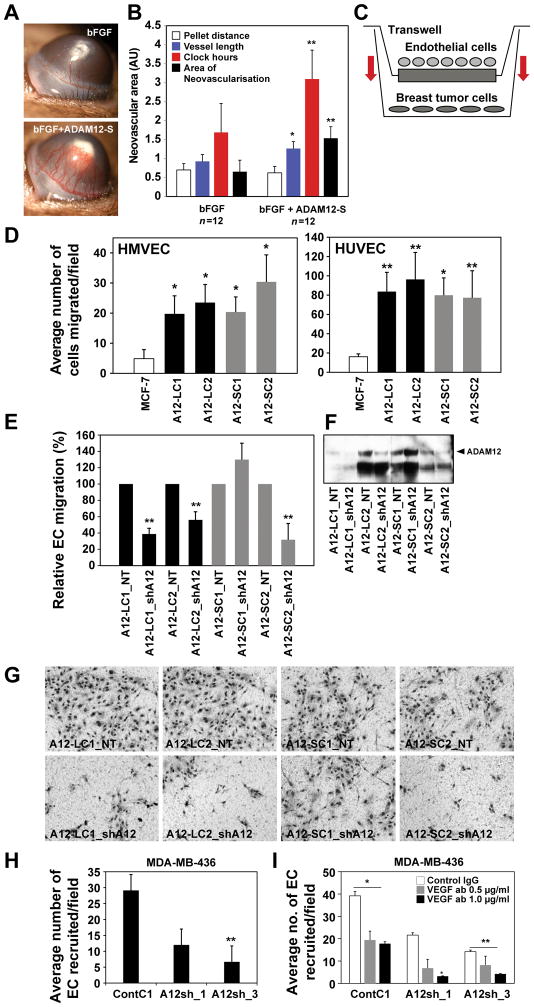
To determine whether ADAM12 regulates normal neovascularization in vivo, we tested recombinant human ADAM12-S in the corneal pocket angiogenesis assay. Slow release methylcellulose pellets containing active rADAM12-S (~80 ng) were surgically implanted into the mouse corneal pocket approximately 1 mm away from the limbus. Recombinant ADAM12-S (or furin control; ~80 ng/pellet) alone had no effect on corneal neovascularization (data not shown). Therefore, we investigated whether ADAM12 may instead potentiate an angiogenic factor-induced neovascular response. ADAM12-S in combination with low levels of bFGF (~30 ng/pellet) generated a significantly (2-fold) higher angiogenic response in the cornea as compared to bFGF alone (Fig. 3A, B). Vessel length, clock hours and total area of neovascularization were significantly higher in the rADAM12-S treated cohort (Fig. 3B). These data suggest that ADAM12 may potentiate growth factor-initiated and not basal angiogenesis in vivo.
Figure 3. ADAM12 induces angiogenesis.
In the mouse corneal pocket assay, rADAM12-S in combination with low levels of bFGF (A;bottom) generated a 2-fold higher angiogenic response in the cornea compared to bFGF alone. Vessel length, clock hours and total area of neovascularization are indicated (B;n=12/treatment, *P<0.05, **P<0.005). In co-culture studies (C), EC migrated at a faster rate towards ADAM12-L (black) and ADAM12–S (grey) expressing BTC compared to MCF-7 (white;D). EC recruitment was reduced when ADAM12 was silenced (E–G). EC recruitment was significantly reduced in MDA-MB-436 after ADAM12-silencing compared to control shRNA (H; 3 independent experiments, one way ANOVA; *P<0.05, **P<0.001). Combined targeting of ADAM12 and VEGF has a synergistic inhibitory effect on EC recruitment (I). MDA-MB-436 ContC1, A12sh_1 and A12sh_3 were treated with control IgG (1 μg/ml) or VEGF-neutralizing antibodies (0.5 μg/ml, 1.0 μg/ml) in the EC recruitment assay. EC recruitment was reduced after VEGF antibody treatment by ~50% in ContC1 cells, whereas ADAM12-silenced cells A12sh_1 and A12_sh3 displayed a ~40–80% further reduction compared to control IgG-treated conditions (I; results average of 2 independent experiments, *P<0.05, **P<0.0001).
ADAM12-expressing breast tumor cells recruit endothelial cells
Based on our observation that ADAM12-expressing breast tumors have a significantly higher microvessel density (Fig. 1G, H), we next investigated the cellular basis for this ADAM12-dependent phenotype. Aggressively metastatic tumor cells have previously been shown to recruit EC at higher rates than less metastatic cells (28). When ADAM12-L or ADAM12-S overexpressing MCF-7 cells were placed in the bottom well of a transwell they recruited HMVEC or HUVEC across the transwell filter at significantly higher rates as compared to the WT MCF-7 (Fig. 3C, D; *p<0.01, **p<0.001). The stimulation of EC recruitment was observed for multiple ADAM12-L-and ADAM12-S-expressing clones (Fig. 3D). Treatment with GM6001, a broad spectrum metalloprotease inhibitor, had a slight inhibitory effect on ADAM12-S-expressing clones but no effect on ADAM12-L-expressing cells, suggesting that ADAM12 catalytic function may not be absolutely essential for EC recruitment (Suppl Fig. 2A). However, given that GM6001 is not a potent inhibitor of ADAM12 activity (26), and in the absence of a specific inhibitor of ADAM12, we downregulated ADAM12 via shRNA and this resulted in a proportional decrease (50–75%) of EC recruitment for both ADAM12-L and ADAM12-S-expressing BTC (breast tumor cells), (Fig. 3E–G) indicating that the observed effect on EC recruitment is specific for ADAM12. The inhibition of EC recruitment appeared to be directly proportional to ADAM12 levels (Fig. 3F). For the A12-SC1 clone which did not have an appreciable ADAM12 reduction, we did not observe an inhibition of EC recruitment (Fig. 3 E–G). Next, we determined whether ADAM12’s effect could be generalized across breast tumor cell lines (BTC). Expression of ADAM12-L isoform in T47-D and MDA-MB-231 also resulted in increased EC recruitment (Suppl. Fig. 2B). Similarly, downregulation of high endogenous ADAM12 in the triple negative BTC MDA-MD-436 resulted in a significant (70–85%) reduction of EC recruitment by these cells (Fig. 3 H). Since VEGF is known to contribute to EC migration, we next asked whether VEGF blockade could further inhibit EC recruitment. Combined targeting of ADAM12 and VEGF had a synergistic inhibitory effect on EC recruitment (Fig. 3I). While EC migration was suppressed ~50% in ContC1 cells treated with VEGF antibodies, ADAM12 silenced MDA-MB-436 cells A12sh_1 and A12sh_3 had ~40–80% further reduction in EC recruitment when treated with VEGF-neutralizing antibodies. These findings suggest that upregulated ADAM12 in the tumor microenvironment potentiates EC recruitment towards the tumor. Taken together, ADAM12’s effect on the stimulation of EC migration, tube-formation and recruitment indicates its potential to regulate tumor angiogenesis in vivo.
ADAM12 regulates angiogenesis via the differential expression of pro- and anti-angiogenic factors
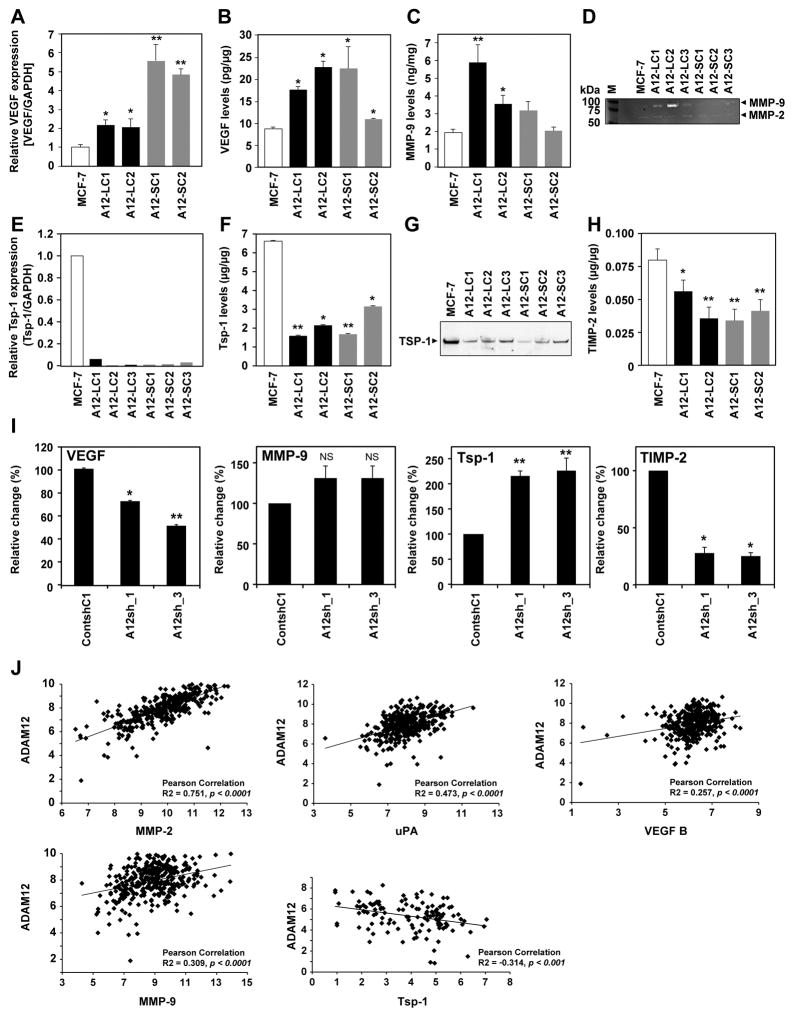
To begin to elucidate the mechanism(s) by which ADAM12 in tumor cells may stimulate tumor angiogenesis or influence de novo neovascularization in the cornea, we utilized an angiogenesis PCR array to profile the expression of key genes involved in modulating the biological processes of angiogenesis. We compared the angiogenic profile of ADAM12-expressing MCF-7 and WT MCF-7 and identified 14 genes that were selectively upregulated or downregulated (>3-fold) in ADAM12-expressing cells (Suppl. Table II).
Validation of the differentially expressed genes via Realtime RT-PCR, ELISA and immunoblot approaches confirmed that ADAM12 expression correlates with significantly increased levels of the pro-angiogenic factors VEGF and MMP-9, whereas anti-angiogenic factors such as TSP1 and TIMP-2 were downregulated (Suppl. Table II, Fig. 4). VEGF mRNA was upregulated between 2–6-fold in ADAM12 clones (Fig. 4A; *P<0.05). Similarly, VEGF protein was 1.5–2-fold higher in ADAM12-expressing cells (Fig. 4B; *P<0.05). MMP-9 protein levels were significantly higher in ADAM12-L-expressing cells (Fig. 4C; *P<0.05). MMP-9 mRNA was upregulated 4–8-fold in ADAM12-L-expressing clones but not in ADAM12-S-expressing cells (Suppl Fig.3A). Increased MMP-9 activity was also detected in CM from ADAM12 clones compared to WT MCF-7 (Fig. 4D), whereas MMP-2 activity remained unchanged (Fig. 4D). Downregulation of ADAM12 in these cells resulted in a ~30–80% reduction in VEGF and 10–40% reduction in MMP-9 respectively (Suppl. Fig. 3B, C), indicating that the upregulation of proangiogenic factors in these cells was a direct result of ADAM12. Although we did identify increased mRNA of several of the angiogenic/inflammatory factors originally identified in the array (prokineticin-2, IL-6, IL-8, MCP-1, Ang-1 and uPA; Suppl. Table II), protein levels of these factors were not upregulated in ADAM12-expressing BTC and therefore were not pursued.
Figure 4. Differential expression of angiogenic regulators in ADAM12-expressing breast tumor cells.
ADAM12-L-expressing tumor cells (black bars) had significantly higher VEGF mRNA and protein (A,B) and MMP-9 protein level and activity (C,D) compared to MCF-7 (white bars, *P≤0.06, **P<0.001). ADAM12-S-expressing breast tumor cells (grey bars) had higher VEGF levels; MMP-9 levels were only slightly increased (C,D). Thrombospondin-1 mRNA (E) and protein (F,G) and TIMP-2 (H) was significantly downregulated in ADAM12-expressing BTC compared to MCF-7 (*P<0.05, **P<0.005). Downregulation of ADAM12 in MDA-MB-436 cells resulted in reduced VEGF and increased TSP1, although, MMP-9 did not change and TIMP-2 expression decreased (I;*P<0.05, **P<0.001). Realtime RT-PCR and ELISA results are an average of 2 independent experiments. Correlation analysis of ADAM12 mRNA with angiogenic factors in a set of human breast tumors (Affymetrix® Gene Profiling Array cGMPU133P2) from publically available datasets indicated a positive correlation of ADAM12 with MMP-2, uPA, VEGF and MMP-9 and a negative correlation with TSP1 (J).
In contrast, inhibitors of neovascularization such as TSP1 and TIMP-2 were significantly downregulated in ADAM12-expressing cells (Suppl. Table II, Fig. 4E–H). This effect was specific since downregulation of ADAM12 in these cells resulted in increase in both TSP1 levels and TIMP-2 levels (Suppl. Fig. 3D, E). While transcript levels of anti-angiogenic factors such as TIMP-1, Platelet factor 4 and endostatin show a >2-fold decrease in the angiogenesis array (Suppl. Table II), protein levels of these candidates did not change in response to ADAM12 expression (data not shown) and therefore, were not pursued. ADAM12 overexpression in T47-D also resulted in the upregulation of VEGF and downregulation of TSP1 and TIMP-2, although MMP-9 levels were undetectable in this cell line (Suppl. Fig. 4). Consistent with these findings, downregulation of endogenous ADAM12 in MDA-MB-436 (Suppl. Fig. 5 A, B) resulted in a 30–50% decrease in VEGF whereas MMP-9 levels remained unchanged (Fig. 4I). However, MDA-MB-436 cells express very low endogenous MMP-9. TSP1 levels were significantly upregulated (2-fold) in response to ADAM12 silencing in MDA-MB-436 cells but TIMP-2 levels did not recover in response to ADAM12 downregulation (Fig. 4I). As expected, proliferation and migration rates of ADAM12-silenced MDA436 cells were significantly reduced (Suppl. Fig. 5C, D). Taken together, these data suggest that differential expression of the pro- and anti-angiogenic factors may be specifically regulated by ADAM12 in a variety of BTC and that, in turn, these factors may stimulate tumor angiogenesis within the tumor microenvironment.
ADAM12 mRNA levels are associated with a pro-angiogenic tumor phenotype
To investigate the physiological relevance of these findings, we next analyzed the correlation of ADAM12 mRNA expression with key angiogenic regulators identified in our study in human breast cancer tissues. Analysis of publically available microarray data of a cohort of ~300 breast cancer patient samples indicated that ADAM12 mRNA levels were positively associated with proangiogenic factors such as VEGF, MMP-9, MMP-2 and uPA (urokinase-type plasminogen activator) while anti-angiogenic factors such as TSP1, appeared to negatively associate with ADAM12 in breast tumor tissues (Fig. 4J, Suppl. Fig. 6). These data indicate that ADAM12 correlates with a pro-angiogenic phenotype in breast tumor tissues.
ADAM12 expression correlates with STAT3 activation in breast tumor cells
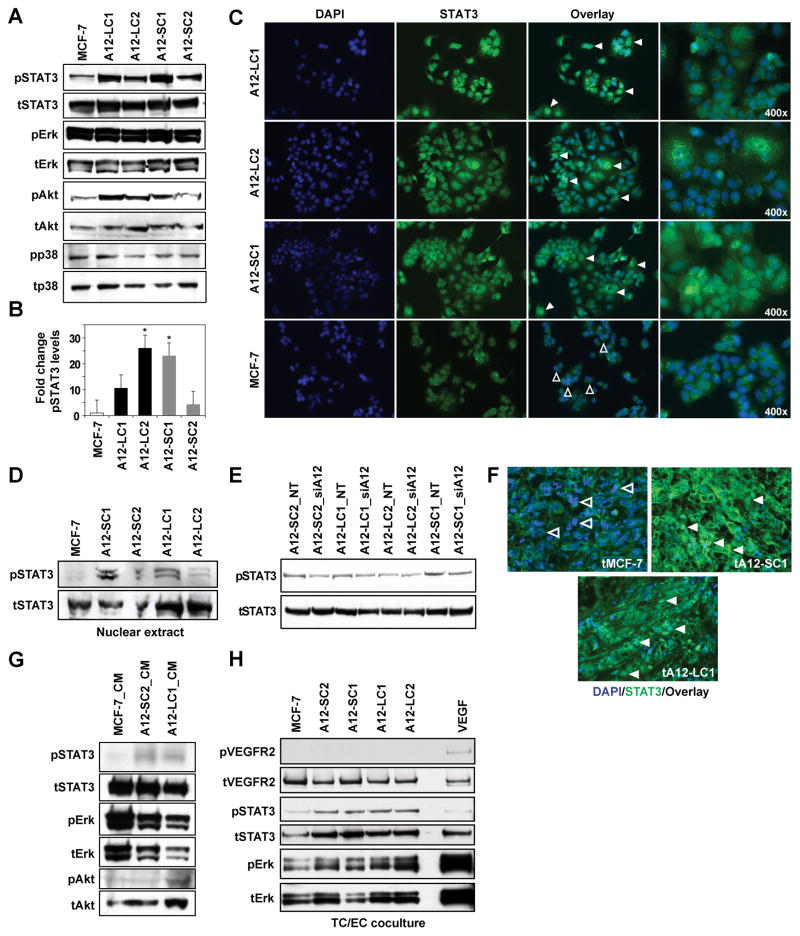
The transcriptional activator STAT3 is now recognized to regulate both physiological and pathological angiogenic processes (29–31). Therefore, we asked whether the ADAM12-mediated proangiogenic phenotype could be STAT3-driven. ADAM12-expressing MCF-7 clones had markedly higher levels of phospho-STAT3 compared to WT MCF-7 cells (Fig. 5A, B). Phospho-Erk and phospho-p38 levels remained unchanged, however, phospho-Akt levels were also upregulated (Fig 5A). Immunocytostaining and immunoblots of nuclear extracts indicated that ADAM12 clones had increased nuclear localization of STAT3 compared to MCF-7 cells (Fig. 5C, D) and downregulation of ADAM12 resulted in a concomitant decrease in pSTAT3 levels (Fig. 5E). ADAM12-expressing T47-D as well as MDA-MB-231 cells also had similar upregulation in pSTAT3 levels (Suppl. Fig 7A, B), although pErk remained unchanged. Consistent with these findings, ADAM12-expressing tumors have increased STAT3 nuclear localization compared to WT MCF-7 tumors (Fig. 5F). Therefore, our data suggest that the pro-angiogenic function of ADAM12-expressing BTC may be mediated via increased STAT3 activation.
Figure 5. STAT3/Akt signaling is activated in ADAM12-expressing breast tumor cells.
PhosphoSTAT3 and pAkt is higher in ADAM12 clones compared to MCF-7 (A). Total protein served as loading control for each phosphoprotein analyzed. Graph depicts fold-change in pSTAT3 (B;ImageJ; ratio pSTAT3/totalSTAT3; mean of 3 blots, *P<0.05 ). STAT3 (green) localized to the nucleus (DAPI) in ADAM12 clones (C, solid arrows), whereas there was no overlap in MCF-7 cells (open arrows). Nuclear extracts from ADAM12 clones have increased pSTAT3 compared to MCF-7 (D). Silencing of ADAM12 resulted in downregulation of pSTAT3 (E). Enhanced nuclear localization of STAT3 was also detected in representative ADAM12-L and ADAM12-S-expressing tumors (solid arrows) but not in WT MCF-7 tumors (F, open arrows). Treatment of WT MCF-7 cells with CM from ADAM12-L or –S clones resulted in increased pSTAT3 and pAkt (G). Co-culture of EC with ADAM12-expressing clones upregulated pSTAT3 and pErk levels in HMVEC (H). EC treated with VEGF served as positive control. Immunoblots shown are representative of 3 independent experiments.
To determine whether ADAM12 function in BTC may upregulate STAT3 activation in surrounding tumor or endothelial cells, MCF-7 cells were treated with CM from WT MCF-7 or ADAM12-L- or ADAM12-S-expressing clones respectively, and the activation status of STAT3, Erk and Akt was determined. Treatment of WT MCF-7 cells with CM from ADAM12-expressing cells resulted in an upregulation of pSTAT3 and pAkt (Fig. 5G). Since VEGF levels were ~2-fold upregulated in ADAM12-expressing tumor cells (Fig. 4A, B), we determined whether pVEGFR2 levels are upregulated in EC under co-culture conditions. While pVEGFR2 levels were not upregulated in HMVECs cocultured with BTC (Fig. 5H), interestingly, pSTAT3 and pErk levels were significantly upregulated in EC cocultured with ADAM12-expressing tumor cells as compared to those cultured with WT MCF-7 cells (Fig. 5H). Our results suggest that ADAM12 expression in tumors results in increased activation of STAT3 in surrounding tumor cells as well as an increased activation of pSTAT3 and pErk in the endothelium within the tumor microenvironment.
ADAM12 stimulates EC recruitment in a STAT3-dependent manner
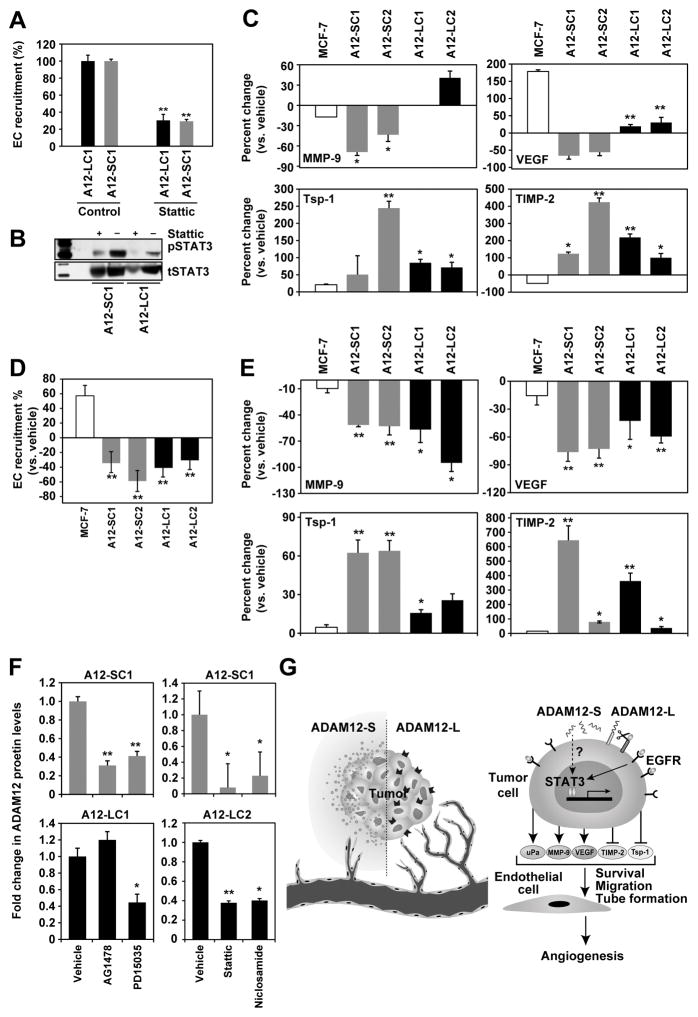
To determine whether ADAM12’s proangiogenic effect is mediated via STAT3 we asked whether STAT3 inhibition has an effect on the expression of proangiogenic factors or EC recruitment. Inhibition of STAT3 (Stattic; 5μm) resulted in ~70% inhibition of EC recruitment by ADAM12-expressing BTC (Fig. 6A). Downregulation of pSTAT3 was confirmed upon Stattic treatment of ADAM12-expressing tumor cells (Fig. 6B). In agreement with these data, STAT3 mRNA silencing in ADAM12-L and –S clones also resulted in a significant reduction of EC recruitment (Suppl. Fig 8A, B). Inhibition of STAT3 lead to a 40–70% decrease in MMP-9 levels in ADAM12-S clones although the effect was not seen in ADAM12-L-expressing cells. Similarly, VEGF in ADAM12-expressing BTC was significantly downregulated in response to STAT3 inhibition (Fig.6 C). Conversely, STAT3 inhibition resulted in a significant increase in the anti-angiogenic factors TSP1 and TIMP-2 levels (Fig. 6 C). PhosphoSTAT3 levels were downregulated in response to STAT3 inhibitor treatment, although pErk levels remained unchanged (Suppl. Fig. 7C). Taken together, our data suggest that the proangiogenic effects of ADAM12 may be mediated via an upregulation of STAT3 activation in tumor cells which in turn creates a positive angiogenic balance within the tumor stimulating EC recruitment and tube formation. Tumor-associated ADAM12 may also activate the STAT3/ERK cascade in the naïve tumor-associated endothelium or surrounding epithelium proximal to the tumor.
Figure 6. Targeting STAT3 and EGFR abrogates the proangiogenic phenotype of ADAM12-expressing BTC.
Treatment with STAT3 inhibitor (Stattic;5μM) resulted in downregulation of EC recruitment by these cells (A) and downregulation of MMP-9 and VEGF and upregulation of TSP1 and TIMP-2 in ADAM12 clones but not in MCF-7 (C). ADAM12-L (black), ADAM12-S (grey) and MCF-7 (white). Reduction of pSTAT3 after Stattic treatment (B). Total STAT3 served as the loading control. EGFR inhibition (AG1478;10μM) in ADAM12 clones lead to abrogation of EC recruitment by these cells (D), and downregulation of MMP-9 and VEGF and upregulation of TSP1 and TIMP-2 versus vehicle-treated cells (E). MCF-7 levels did not change (E). Targeting both EGFR (AG1478 and PD15035) and STAT3 (Static and Niclosamide) in ADAM12-expressing clones resulted in a downregulation of ADAM12 protein (F; versus vehicle-treated cells). ELISA results are the mean of 2 independent experiments. Working model for ADAM12-mediated regulation of tumor angiogenesis (G). Upregulation of ADAM12 in breast tumors leads to increased transactivation of EGFR which in turn activates STAT3 signaling and a pro-angiogenic shift within the tumor milieu. These changes in the tumor microenvironment result in robust EC recruitment and tube-formation and ultimately enhanced tumor angiogenesis.
ADAM12-induced STAT3 activation is mediated via EGFR
We have previously reported that in BTC, ADAM12 upregulates EGFR and MAPK activation (15). In a variety of tumors, membrane-associated ADAM12-L is a sheddase for ligands such as HB-EGF (11), EGF, betacellulin and amphiregulin that in turn can activate EGFR in an autocrine or paracrine manner within the tumors (32). ADAM12-L is the protease primarily responsible for ligand release and activation of EGFR in triple negative breast cancers (5). Since EGFR signaling is known to regulate STAT3 activation in tumor cells (33–35), we asked whether EGFR inhibition or downregulation in our breast tumor model would attenuate ADAM12-mediated proangiogenic effects. EGFR inhibition via AG1478 (10 μM) or protein downregulation via silencing resulted in a significant reduction of EC recruitment towards ADAM12-expressing BTC (Fig. 6D, Suppl. Fig 8C, D). In addition, AG1478 treatment also downregulated proangiogenic factors VEGF and MMP-9 and upregulated antiangiogenic factors TSP1 and TIMP-2 (Fig. 6E). Finally, EGFR inhibition also resulted in downregulation of pSTAT3 and pErk levels in ADAM12-expressing BTC (Suppl. Fig. 7D), suggesting that EGFR activation is an upstream event of STAT3 signaling in these cells. Finally, we investigated whether EGFR and/or STAT3 inhibition would downregulate ADAM12 expression itself in BTC. EGFR or STAT3 inhibition resulted in significant downregulation of both ADAM12 isoforms in these cells (Fig. 6F). These data suggest that a feed-forward loop may exist between ADAM12 and STAT3/EGFR in tumor and EC in the tumor microenvironment. Taken together, our results suggest that increased EGFR activation in response to ADAM12 expression in tumor cells leads to activation of EGFR/STAT3/Akt signaling pathways, which in turn shifts the balance towards angiogenesis within the tumor microenvironment (Fig. 6G).
Discussion
ADAM12 has been reported to be upregulated in tumor-associated vasculature in ovarian (24,25), breast (26) and skin (8) tumors. However, whether ADAM12 promotes or contributes to tumor angiogenesis is unclear. In the current study, we have found that MCF-7 tumors expressing both isoforms of ADAM12 had significantly higher MVD and that tumors that have undergone the switch to the angiogenic phenotype displayed higher ADAM12 levels. In addition, while ADAM12 is low or absent in quiescent EC, ADAM12 mRNA and protein levels are significantly upregulated in the activated endothelium. Interestingly, VEGF treatment of EC resulted in ADAM12 localization to the ruffled edges or invadopodia like structures (Fig. 2). Previous reports have described a role for ADAM12 in invadopodia formation in tumor cells resulting in increased cell invasion under hypoxic conditions (36,37). Therefore, it is feasible that VEGF treatment of EC induces localization of ADAM12 within invadopodia thereby stimulating VEGF-induced EC migration. Consistent with these observations, we found that the transient expression of ADAM12-L or ADAM12-S in two distinct EC lines resulted in increased cell migration and tube-formation suggesting that these EC properties are affected by ADAM12 functions. Finally, when we tested recombinant ADAM12-S in a corneal micropocket assay, it promoted robust angiogenesis only in the presence of low levels of bFGF (Fig. 3), suggesting that ADAM12 contributes to growth-factor initiated but not basal angiogenesis. ADAM12 levels were also upregulated in EC in response to cytokines such as TNFα and TGFβ. The angiogenic actions of TNFα are not clear, given that it is generally considered proangiogenic in vivo but antiangiogenic in vitro. Interestingly, while continuous TNFα treatment inhibits EC proliferation and migration, intermittent treatment does indeed stimulate tube-formation and induces a tip cell phenotype in EC (38) via upregulation of Notch signaling in a NFKβ-dependent manner. Notch signaling has also been reported to be mediated via ADAM12 function (36). Therefore, it is feasible that TNFα stimulation of the endothelium results in increased Notch signaling as well as ADAM12-induced tubulogenesis in our model.
A direct contribution for any ADAM to tumor angiogenesis has not yet been reported, although several ADAMs including ADAM10 (39,40), ADAM15 (41,42), ADAM17 (43,44) and ADAM33 (45) have been implicated in pathological angiogenesis related to rheumatoid arthritis, atherosclerosis and oxygen-induced retinopathy. Transient ADAM12-L overexpression in mouse and human EC resulted in significantly increased shedding of ligands such as KitL1, VE-Cadherin, and Tie-2 in vitro (26), suggesting a contribution to vascular development and/or maintenance. However, how such shedding events may affect the endothelium or the mechanism(s) that underlie ADAM12-mediated tumor angiogenesis remain unknown. ADAM12 function promotes tumor proliferation (3,13,14) as well as local and distant tumor metastasis (3,4,46). Hints that ADAM12 may directly contribute to the metastatic process are beginning to emerge. In an animal model of Lewis Lung carcinoma, ADAM12-cleaved ephrin-A1 enhanced vascular permeability and increased soluble ephrin-A1 levels in the serum, which in turn facilitated tumor cell recruitment to the lungs and increased lung metastasis (46). It is widely accepted that both tumor progression and metastasis are driven by angiogenesis. Therefore, we sought to investigate whether increased ADAM12 expression and function in tumor cells may facilitate tumor-associated angiogenesis in a paracrine manner. Highly metastatic cells are known to recruit endothelial cells to facilitate tumor angiogenesis and more efficient metastatic colonization (28,47).
In this study, increased ADAM12 in BTC (endogenous and stable overexpression) resulted in significantly higher rates of EC recruitment. This effect was observed across different types of BTC and was abrogated when ADAM12 was silenced and partially inhibited when the cells were treated with a broad-spectrum metalloprotease inhibitor, indicating that these effects were directly regulated by ADAM12. Cross-talk between tumor cells and their microenvironment may be facilitated by a variety of soluble factors and affect processes such as tumor cell survival, angiogenesis, vascular permeability and metastasis. To investigate the soluble factors which may stimulate ADAM12-mediated EC recruitment, we used a PCR-based angiogenesis array. Pro-angiogenic factors such as VEGF and MMP-9 were upregulated in ADAM12-expressing BTC compared to WT MCF-7 cells, whereas antiangiogenic factors such as TSP1 and TIMP-2 were significantly downregulated. As with EC recruitment, the dysregulation of angiogenic factors was the direct effect of ADAM12. Moreover, analysis of publicly available microarray datasets confirmed a positive correlation between high ADAM12 and MMP-9 and VEGF mRNA expression and an inverse correlation with TSP1 in a cohort of human breast tumor tissues. Interestingly, downregulation of ADAM12 combined with VEGF targeting resulted in a synergistic reduction of EC recruitment in breast tumor cells. Therefore, ADAM12 levels in tumor cells in culture, in an orthotopic breast tumor model as well as human breast tumor tissues all appear to correlate with increased proangiogenic factors and reduced antiangiogenic factors. Taken together, these data suggest that ADAM12 in tumor cells may facilitate a proangiogenic tumor microenvironment which leads to enhanced tumor growth and more efficient metastasis.
We then investigated the mechanism(s) by which ADAM12 may regulate these proangiogenic effects. In ADAM12-expressing BTC phospho-Erk and phospho-p38 levels remained unchanged, however, phospho-STAT3 and phospho-Akt levels were increased compared to MCF-7 cells (Fig. 5). This was accompanied by increased nuclear localization of STAT3 in ADAM12-expressing cells and tumors. Silencing of ADAM12 reversed the STAT3 activation considerably. Co-culture of ADAM12-expressing cells with WT MCF-7 was sufficient to activate pSTAT3 and pAkt in the latter. We also observed increased activation of pSTAT3 and pErk when ADAM12-expressing BTC were co-cultured with EC, but this effect was not observed for MCF-7. These data suggest that ADAM12 in BTC can activate tumor or endothelial cells within the tumor microenvironment. Bioinformatics analysis has identified STAT3-regulated signaling pathways in more aggressive basal-like breast cancers (48). STAT3 is a direct transcriptional activator of VEGF (30,31), and persistently activated STAT3 can transform human mammary epithelial cells via significantly increased MMP-9 activity (49). We have observed a significant increase in both VEGF and MMP-9 levels in ADAM12-expressing BTC. Therefore, increased activation of STAT3 in ADAM12-expressing cells could result in upregulated angiogenic factors and a proangiogenic tumor microenvironment. Consistent with this hypothesis, we found that inhibition of STAT3 in these cells resulted in significant inhibition of EC recruitment as well as reduction in VEGF and MMP-9 levels and increased TSP1 and TIMP-2 levels, suggesting that the proangiogenic phenotype observed in ADAM12-expressing BTC was directly mediated via STAT3 signaling.
In tumors, stimulation by cytokines such as IL-6, IL-10 or MCP-1 or growth factors may be involved in persistent STAT3 activation (29). In the current study, although the angiogenesis array detected a significant upregulation of IL-6, MCP-1 and CXCL-10 mRNA levels in ADAM12-expressing BTC, protein levels of these factors seemed to be unchanged compared to WT MCF-7 cells (Suppl. Table II). Transactivated EGFR has recently been shown to induce activation of STAT3 in a cardiac hypertrophy (35), similarly, elevated EGFR expression or phosphorylation has been reported to upregulate VEGF levels and potentiate angiogenesis via the activation of STAT3 (33,34). We and others have previously reported that ADAM12-L expression in tumor cells results in increased EGFR expression as well as phosphorylation (3,13,15). We therefore asked whether EGFR signaling may lie upstream of STAT3 activation and the proangiogenic phenotype of ADAM12-expressing BTC. Indeed, EGFR inhibition in ADAM12-expressing BTC resulted in complete abrogation of EC recruitment and a significant downregulation of VEGF and MMP-9 levels and upregulation of TSP1 and TIMP-2. We also found that EGFR inhibition downregulates pSTAT3 levels thereby confirming that enhanced EGFR signaling in ADAM12-expressing BTC directly increases STAT3 activation and that EGFR lies upstream of STAT3 in this signaling cascade. Notably, EGFR (and to a certain extent STAT3) inhibition also resulted in decreased expression of ADAM12, indicating that a feed-forward loop may exist in breast tumor cells, whereby ADAM12 function upregulates EGFR expression and/or activation which in turn increases STAT3 phosphorylation and its downstream proangiogenic influence in the tumor microenvironment. However, once activated EGFR and STAT3 may also drive increased expression of ADAM12 to maintain the malignant signaling cascade.
While membrane-associated ADAM12-L function could certainly activate downstream EGFR and STAT3 signaling via shedding of a variety of ligands e.g., amphiregulin, EGF and HB-EGF in BTC (4,6,17,19), it is unclear how the secreted ADAM12-S isoform activates pSTAT3. While ADAM12-S in BTC did result in increased phosphorylated STAT3 levels, this may or may not occur via EGFR. ADAM12-S is not known to shed cell-surface EGF ligands or activate EGFR (15), however, EGFR inhibition in ADAM12-S-expressing tumor cells resulted in a clear reversal of the proangiogenic phenotype. This conflicting finding may be explained by an indirect inhibition of the Akt pathway by the EGFR inhibtiors in these cells. Alternately, ADAM12-S is also known to cleave IGFBP-3 and -5 (50), therefore increased bioavailability of free IGF could result in increased activation of IGF-1R and this could, in turn, activate STAT3 signaling (51). Finally, we cannot rule out the possibility that ADAM12’s proangiogenic effect may be in part due to shedding of ligands such as Tie-2, Flt-1 and VCAM in the tumor endothelium. To our knowledge, this report is the first to establish a link between elevated ADAM12 levels in the tumor epithelium and a concomitant proangiogenic effect on the tumor microenvironment. Increased ADAM12 expression and function has been reported in the major breast tumor subtypes including triple negative (52), ER+ and Her2-positive breast tumors (13,17,53) suggesting that ADAM12 may contribute to the development of aggressive and metastatic disease regardless of the tumor subtype. As outlined in the current study, ADAM12 also regulates tumor angiogenesis. Combined targeting of this protease with standard therapies holds promise for the treatment of all types of breast cancer.
Supplementary Material
Implications.
These novel findings suggest that ADAM12 regulates EC function and facilitates a proangiogenic microenvironment in a STAT3-dependent manner. A combined approach of targeting ADAM12 and STAT3 signaling in breast tumors may represent a promising strategy to inhibit tumor neovascularization.
Acknowledgments
This work was supported by The Breast Cancer Research Foundation and NIH R01CA185530. The authors thank Ulla M. Wewer (University of Copenhagen, Copenhagen, Denmark) for providing the expression plasmids for ADAM12 isoforms. We thank Kristin Johnson for assistance with the figures.
Abbreviations
- ADAM12
A Disintegrin and Metalloprotease
References
- 1.Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. The Journal of biological chemistry. 2004;279:51323–30. doi: 10.1074/jbc.M409565200. [DOI] [PubMed] [Google Scholar]
- 2.Frohlich C, Nehammer C, Albrechtsen R, Kronqvist P, Kveiborg M, Sehara-Fujisawa A, et al. ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression. Molecular cancer research : MCR. 2011;9:1449–61. doi: 10.1158/1541-7786.MCR-11-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy R, Rodig S, Bielenberg D, Zurakowski D, Moses MA. ADAM12 transmembrane and secreted isoforms promote breast tumor growth: a distinct role for ADAM12-S protein in tumor metastasis. The Journal of biological chemistry. 2011;286:20758–68. doi: 10.1074/jbc.M110.216036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kveiborg M, Frohlich C, Albrechtsen R, Tischler V, Dietrich N, Holck P, et al. A role for ADAM12 in breast tumor progression and stromal cell apoptosis. Cancer research. 2005;65:4754–61. doi: 10.1158/0008-5472.CAN-05-0262. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Duhachek-Muggy S, Dubnicka S, Zolkiewska A. Metalloproteinase-disintegrin ADAM12 is associated with a breast tumor-initiating cell phenotype. Breast cancer research and treatment. 2013;139:691–703. doi: 10.1007/s10549-013-2602-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peduto L, Reuter VE, Sehara-Fujisawa A, Shaffer DR, Scher HI, Blobel CP. ADAM12 is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression. Oncogene. 2006;25:5462–6. doi: 10.1038/sj.onc.1209536. [DOI] [PubMed] [Google Scholar]
- 7.Cheon DJ, Li AJ, Beach JA, Walts AE, Tran H, Lester J, et al. ADAM12 is a prognostic factor associated with an aggressive molecular subtype of high-grade serous ovarian carcinoma. Carcinogenesis. 2015;36:739–47. doi: 10.1093/carcin/bgv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao VH, Vogel K, Yanagida JK, Marwaha N, Kandel A, Trempus C, et al. Erbb2 up-regulation of ADAM12 expression accelerates skin cancer progression. Molecular carcinogenesis. 2015;54:1026–36. doi: 10.1002/mc.22171. [DOI] [PubMed] [Google Scholar]
- 9.Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Rocken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol. 2005;26:17–24. [PubMed] [Google Scholar]
- 10.Mino N, Miyahara R, Nakayama E, Takahashi T, Takahashi A, Iwakiri S, et al. A disintegrin and metalloprotease 12 (ADAM12) is a prognostic factor in resected pathological stage I lung adenocarcinoma. Journal of surgical oncology. 2009;100:267–72. doi: 10.1002/jso.21313. [DOI] [PubMed] [Google Scholar]
- 11.Kodama T, Ikeda E, Okada A, Ohtsuka T, Shimoda M, Shiomi T, et al. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. The American journal of pathology. 2004;165:1743–53. doi: 10.1016/S0002-9440(10)63429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilpin BJ, Loechel F, Mattei MG, Engvall E, Albrechtsen R, Wewer UM. A novel, secreted form of human ADAM 12 (meltrin alpha) provokes myogenesis in vivo. The Journal of biological chemistry. 1998;273:157–66. doi: 10.1074/jbc.273.1.157. [DOI] [PubMed] [Google Scholar]
- 13.Rao VH, Kandel A, Lynch D, Pena Z, Marwaha N, Deng C, et al. A positive feedback loop between HER2 and ADAM12 in human head and neck cancer cells increases migration and invasion. Oncogene. 2012;31:2888–98. doi: 10.1038/onc.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georges S, Chesneau J, Hervouet S, Taurelle J, Gouin F, Redini F, et al. A Disintegrin And Metalloproteinase 12 produced by tumour cells accelerates osteosarcoma tumour progression and associated osteolysis. European journal of cancer. 2013;49:2253–63. doi: 10.1016/j.ejca.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Roy R, Moses MA. ADAM12 induces estrogen-independence in breast cancer cells. Breast cancer research and treatment. 2012;131:731–41. doi: 10.1007/s10549-011-1431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy R, Zurakowski D, Pories S, Moss ML, Moses MA. Potential of fluorescent metalloproteinase substrates for cancer detection. Clinical biochemistry. 2011;44:1434–9. doi: 10.1016/j.clinbiochem.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma B, Ma Q, Jin C, Wang X, Zhang G, Zhang H, et al. ADAM12 expression predicts clinical outcome in estrogen receptor-positive breast cancer. International journal of clinical and experimental pathology. 2015;8:13279–83. [PMC free article] [PubMed] [Google Scholar]
- 18.Shimura T, Dagher A, Sachdev M, Ebi M, Yamada T, Yamada T, et al. Urinary ADAM12 and MMP-9/NGAL complex detect the presence of gastric cancer. Cancer prevention research. 2015;8:240–8. doi: 10.1158/1940-6207.CAPR-14-0229. [DOI] [PubMed] [Google Scholar]
- 19.Frohlich C, Albrechtsen R, Dyrskjot L, Rudkjaer L, Orntoft TF, Wewer UM. Molecular profiling of ADAM12 in human bladder cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:7359–68. doi: 10.1158/1078-0432.CCR-06-1066. [DOI] [PubMed] [Google Scholar]
- 20.Bilgin Dogru E, Dizdar Y, Aksit E, Ural F, Sanli O, Yasasever V. EMMPRIN and ADAM12 in prostate cancer: preliminary results of a prospective study. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:11647–53. doi: 10.1007/s13277-014-2514-8. [DOI] [PubMed] [Google Scholar]
- 21.Shao S, Li Z, Gao W, Yu G, Liu D, Pan F. ADAM-12 as a diagnostic marker for the proliferation, migration and invasion in patients with small cell lung cancer. PloS one. 2014;9:e85936. doi: 10.1371/journal.pone.0085936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pories SE, Zurakowski D, Roy R, Lamb CC, Raza S, Exarhopoulos A, et al. Urinary metalloproteinases: noninvasive biomarkers for breast cancer risk assessment. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:1034–42. doi: 10.1158/1055-9965.EPI-07-0365. [DOI] [PubMed] [Google Scholar]
- 23.Kurisaki T, Masuda A, Sudo K, Sakagami J, Higashiyama S, Matsuda Y, et al. Phenotypic analysis of Meltrin alpha (ADAM12)-deficient mice: involvement of Meltrin alpha in adipogenesis and myogenesis. Molecular and cellular biology. 2003;23:55–61. doi: 10.1128/MCB.23.1.55-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu C, Bonome T, Li Y, Kamat AA, Han LY, Schmandt R, et al. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer research. 2007;67:1757–68. doi: 10.1158/0008-5472.CAN-06-3700. [DOI] [PubMed] [Google Scholar]
- 25.Sasaroli D, Gimotty PA, Pathak HB, Hammond R, Kougioumtzidou E, Katsaros D, et al. Novel surface targets and serum biomarkers from the ovarian cancer vasculature. Cancer biology & therapy. 2011;12:169–80. doi: 10.4161/cbt.12.3.16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frohlich C, Klitgaard M, Noer JB, Kotzsch A, Nehammer C, Kronqvist P, et al. ADAM12 is expressed in the tumour vasculature and mediates ectodomain shedding of several membrane-anchored endothelial proteins. The Biochemical journal. 2013;452:97–109. doi: 10.1042/BJ20121558. [DOI] [PubMed] [Google Scholar]
- 27.Dudley AC, Khan ZA, Shih SC, Kang SY, Zwaans BM, Bischoff J, et al. Calcification of multipotent prostate tumor endothelium. Cancer Cell. 2008;14:201–11. doi: 10.1016/j.ccr.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–4. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 29.Chen Z, Han ZC. STAT3: a critical transcription activator in angiogenesis. Medicinal research reviews. 2008;28:185–200. doi: 10.1002/med.20101. [DOI] [PubMed] [Google Scholar]
- 30.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 31.Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–29. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 32.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. The international journal of biochemistry & cell biology. 2008;40:1685–702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Ranayhossaini DJ, Lu J, Mabus J, Gervais A, Lingham RB, Fursov N. EGF potentiation of VEGF production is cell density dependent in H292 EGFR wild type NSCLC cell line. International journal of molecular sciences. 2014;15:17686–704. doi: 10.3390/ijms151017686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habib AA, Chun SJ, Neel BG, Vartanian T. Increased expression of epidermal growth factor receptor induces sequestration of extracellular signal-related kinases and selective attenuation of specific epidermal growth factor-mediated signal transduction pathways. Molecular cancer research : MCR. 2003;1:219–33. [PubMed] [Google Scholar]
- 35.Li Y, Zhang H, Liao W, Song Y, Ma X, Chen C, et al. Transactivated EGFR mediates alpha(1)-AR-induced STAT3 activation and cardiac hypertrophy. American journal of physiology Heart and circulatory physiology. 2011;301:H1941–51. doi: 10.1152/ajpheart.00338.2011. [DOI] [PubMed] [Google Scholar]
- 36.Diaz B, Yuen A, Iizuka S, Higashiyama S, Courtneidge SA. Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. The Journal of cell biology. 2013;201:279–92. doi: 10.1083/jcb.201209151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albrechtsen R, Stautz D, Sanjay A, Kveiborg M, Wewer UM. Extracellular engagement of ADAM12 induces clusters of invadopodia with localized ectodomain shedding activity. Experimental cell research. 2011;317:195–209. doi: 10.1016/j.yexcr.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, et al. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood. 2008;111:4997–5007. doi: 10.1182/blood-2007-08-108597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isozaki T, Rabquer BJ, Ruth JH, Haines GK, 3rd, Koch AE. ADAM-10 is overexpressed in rheumatoid arthritis synovial tissue and mediates angiogenesis. Arthritis and rheumatism. 2013;65:98–108. doi: 10.1002/art.37755. [DOI] [PubMed] [Google Scholar]
- 40.Glomski K, Monette S, Manova K, De Strooper B, Saftig P, Blobel CP. Deletion of Adam10 in endothelial cells leads to defects in organ-specific vascular structures. Blood. 2011;118:1163–74. doi: 10.1182/blood-2011-04-348557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horiuchi K, Weskamp G, Lum L, Hammes HP, Cai H, Brodie TA, et al. Potential role for ADAM15 in pathological neovascularization in mice. Molecular and cellular biology. 2003;23:5614–24. doi: 10.1128/MCB.23.16.5614-5624.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C, Wu MH, Lee ES, Yuan SY. A disintegrin and metalloproteinase 15 contributes to atherosclerosis by mediating endothelial barrier dysfunction via Src family kinase activity. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:2444–51. doi: 10.1161/ATVBAHA.112.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan D, Takawale A, Shen M, Wang W, Wang X, Basu R, et al. Cardiomyocyte A Disintegrin And Metalloproteinase 17 (ADAM17) Is Essential in Post-Myocardial Infarction Repair by Regulating Angiogenesis. Circulation Heart failure. 2015;8:970–9. doi: 10.1161/CIRCHEARTFAILURE.114.002029. [DOI] [PubMed] [Google Scholar]
- 44.Weskamp G, Mendelson K, Swendeman S, Le Gall S, Ma Y, Lyman S, et al. Pathological neovascularization is reduced by inactivation of ADAM17 in endothelial cells but not in pericytes. Circulation research. 2010;106:932–40. doi: 10.1161/CIRCRESAHA.109.207415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puxeddu I, Pang YY, Harvey A, Haitchi HM, Nicholas B, Yoshisue H, et al. The soluble form of a disintegrin and metalloprotease 33 promotes angiogenesis: implications for airway remodeling in asthma. The Journal of allergy and clinical immunology. 2008;121:1400–6. 6 e1–4. doi: 10.1016/j.jaci.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Ieguchi K, Omori T, Komatsu A, Tomita T, Deguchi A, Maru Y. Ephrin-A1 expression induced by S100A8 is mediated by the toll-like receptor 4. Biochemical and biophysical research communications. 2013;440:623–9. doi: 10.1016/j.bbrc.2013.09.119. [DOI] [PubMed] [Google Scholar]
- 47.Catalano V, Turdo A, Di Franco S, Dieli F, Todaro M, Stassi G. Tumor and its microenvironment: a synergistic interplay. Seminars in cancer biology. 2013;23:522–32. doi: 10.1016/j.semcancer.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Tell RW, Horvath CM. Bioinformatic analysis reveals a pattern of STAT3-associated gene expression specific to basal-like breast cancers in human tumors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12787–92. doi: 10.1073/pnas.1404881111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, Wang Z, Fan Y, Xu Q, Ji W, Tian R, et al. Elevated STAT3 Signaling-Mediated Upregulation of MMP-2/9 Confers Enhanced Invasion Ability in Multidrug-Resistant Breast Cancer Cells. International journal of molecular sciences. 2015;16:24772–90. doi: 10.3390/ijms161024772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loechel F, Fox JW, Murphy G, Albrechtsen R, Wewer UM. ADAM 12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochemical and biophysical research communications. 2000;278:511–5. doi: 10.1006/bbrc.2000.3835. [DOI] [PubMed] [Google Scholar]
- 51.Zong CS, Chan J, Levy DE, Horvath C, Sadowski HB, Wang LH. Mechanism of STAT3 activation by insulin-like growth factor I receptor. The Journal of biological chemistry. 2000;275:15099–105. doi: 10.1074/jbc.M000089200. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Duhachek-Muggy S, Qi Y, Hong Y, Behbod F, Zolkiewska A. An essential role of metalloprotease-disintegrin ADAM12 in triple-negative breast cancer. Breast cancer research and treatment. 2012;135:759–69. doi: 10.1007/s10549-012-2220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narita D, Anghel A, Seclaman E, Ilina R, Cireap N, Ursoniu S. Molecular profiling of ADAM12 gene in breast cancers. Romanian journal of morphology and embryology = Revue roumaine de morphologie et embryologie. 2010;51:669–76. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.