Abstract
Objective
The etiology of knee osteoarthritis (OA), the most common form of arthritis, is complex and may differ by race or ethnicity. In recent years, genetic studies have identified multiple genetic variants associated with OA, but nearly all were conducted in European Caucasian and Asian Americans. Few studies have focused on genetics of knee OA in African Americans.
Methods
We performed a genome-wide association study of radiographic knee OA in 1,217 African Americans from two North American cohort studies: 590 subjects from the Johnston County Osteoarthritis Project and 627 subjects from the Osteoarthritis Initiative. Analyses were conducted in each cohort separately and combined in an inverse variance fixed effects meta-analysis, which were then included in pathway analyses. We additionally tested 12 SNPs robustly associated with OA in European Caucasian populations for association in African Americans.
Results
We identified a genome-wide significant variant in LINC01006 (minor allele frequency=12%, p-value=4.11×10−9) that is less common in European Caucasian populations (minor allele frequency < 3%). Five other independent loci reached suggestive significance (p-value < 1×10−6). In pathway analyses, dorsal/ventral neural tube patterning and iron ion transport pathways were significantly associated with knee OA in African Americans (FDR < 0.05). We found no evidence that previously reported OA susceptibility variants in European Caucasians were associated with knee OA in African Americans.
Conclusion
These results highlight differences in the genetic architecture of knee OA between African American and European Caucasians, which underscores the need to include more diverse populations in OA genetics studies.
Introduction
Osteoarthritis (OA) is a multifactorial joint disease characterized by cartilage degradation and structural changes in the subchondral bone, which often leads to joint pain, activity limitations and physical disability. The etiopathogenesis of OA is complex; family studies indicate that at least part of OA susceptibility is under genetic control (1). The heritability of radiographic knee OA has been estimated in twin studies to be 39% (2). Knee OA is also strongly influenced by environmental factors, such as history of injuries and overweight, making it difficult to identify the genetic mechanisms behind knee OA pathogenesis (3). Identifying genes underlying OA susceptibility is important insofar as the implicated genes may reveal insights into disease pathogenesis and identify potential targets for prevention or therapy. At least a dozen robustly replicated OA susceptibility variants have been identified to date; most of these association studies have been carried out in European Caucasians (4–12).
Studies focused on African Americans are important because African Americans experience a higher prevalence of knee OA (13) than Caucasian Americans and there is disparity in the incidence of total knee arthroplasty for knee OA with higher rates in Caucasian Americans than African Americans (14). It is currently unknown whether these differences are due to biological or sociocultural differences or both. The goals of this report are two-fold: first, to identify SNPs associated with radiographic knee OA in African Americans through a genome-wide association analysis, and second, to determine whether SNPs robustly associated with OA in European Caucasian populations are also associated with radiographic knee OA in African Americans. To address these goals, we performed association analyses in two large population-based studies of knee OA that included substantial numbers of African Americans.
Methods
The analyses presented are based on African American participants of the Johnston County Osteoarthritis Project (JoCo) and the Osteoarthritis Initiative (OAI).
JoCo is an ongoing, community-based cohort study of knee and hip OA in individuals 45 years and older recruited from Johnston County, North Carolina. The original participants were enrolled between 1990 and 1997 (13), with additional participants recruited between 2003 and 2004. Participants were recruited by probability sampling, with oversampling of African Americans, where Caucasians and African Americans represent 68% and 32% of the cohort, respectively. The age of the participants ranged from 45 to 98 years; two-thirds of the participants were women. Additional study details have been previously described (13). JoCo has been continuously approved by institutional review boards (IRB) at the University of North Carolina at Chapel Hill and the Centers for Disease Control and Prevention, and all participants gave informed consent (IRB approval number, 92-0583). A total of 590 African Americans with radiographic readings for knee OA were genotyped and included in this study.
The Osteoarthritis Initiative (OAI) is a longitudinal, natural history study of knee OA in middle aged and older individuals who were either at risk for, or have, symptomatic radiographic knee OA. A total of 4,796 subjects aged 45–79 years were enrolled at four clinical centers (Baltimore, MD; Columbus, OH; Pittsburgh, PA; and Providence, RI), received a baseline evaluation between 2004 and 2006, and were invited back to assess incidence or progression of OA annually for up to eight years. The study was approved by the IRB at each clinical center. All participants provided informed consent. The OAI study and public use of clinical and imaging data used in this study were approved by the committee on Human Research at the University of California, San Francisco (IRB approval number, 10-00532). The current analysis was restricted to 627 African Americans with genotype data and in whom knee radiographic readings, read by consensus, were available from radiographs obtained at the baseline evaluation. Study details and data are publicly available on the OAI web site (http://oai.epi-ucsf.org/datarelease/docs/StudyDesignProtocol.pdf).
Radiographic knee OA cases were defined similarly in both studies as having definite osteophytes and possible joint space narrowing (Kellgren-Lawrence [KL] grade ≥ 2) or total joint replacement in one or both knees. Controls were defined as having in both knees no or doubtful evidence for OA (KL grade = 0 or 1).
Genotyping was performed on the Illumina Infinium 1M-Duo bead array and Illumina Omni-Quad 2.5M array for JoCo and OAI, respectively. Genotypes were called within each study using the Illumina BeadStudio software. Sample level quality control (QC) included removal of samples with below threshold sample call rates (< 0.99 for JoCo and < 0.95 for OAI), samples showing excess evidence for heterozygosity, and samples whose reported gender did not match their sex assignment based on genotype data. Single nucleotide polymorphisms (SNPs) with missing call rate > 0.01 were excluded. We additionally eliminated samples showing evidence for cryptic relatedness based on genetic data and SNPs showing evidence for extreme deviation from Hardy-Weinberg equilibrium (p-value < 1×10−6).
In JoCo and OAI separately, we imputed SNP dosages in all samples based on the 1000 genomes Caucasian and African American reference panel (June 2011 release). Imputation for both studies was conducted using Minimac (http://genome.sph.umich.edu/wiki/Minimac). Following imputation, 8.38 million SNPs were available for analysis in JoCo and OAI.
First, we performed a genome-wide association meta-analysis of the combined JoCo and OAI sample to identify novel SNPs associated with radiographic knee OA in African Americans. Prior to the genetic association analysis, principal components analysis was performed in each study to assess population substructure. Principal components that were significantly associated with the outcome (p-value < 0.05) were used as covariates in association analyses. We tested each SNP for association with radiographic knee OA within JoCo and OAI separately. We used logistic regression, assuming an additive genetic model, with adjustment for age, sex, and principal components. We additionally adjusted for body mass index (BMI) in secondary analyses. Association analyses for the JoCo and OAI were performed with ProbABEL and PLINK, respectively. Beta estimates were then combined across studies weighting the study specific estimates by the inverse of their variances using the METAL software program (15). Based on a combined sample size of 742 cases and 475 controls, we had 80% power to detect odds ratios of 1.51–1.65 for SNPs having minor allele frequencies (MAF) ranging from 0.10–0.50 at genome-wide thresholds for statistical significance (p-value = 5×10−8). Heterogeneity between studies was assessed using Cochran’s Q statistic. We generated LocusZoom plots to visualize and provide genomic context to top meta-analysis findings (p-value < 1×10−6) (16).
Given the limited power to detect associations at individual SNPs, we conducted pathway analyses with MAGENTA (Meta-Analysis Gene-set Enrichment of variant Associations), which can be downloaded from the Broad Institute website (http://www.broadinstitute.org/mpg/magenta/). Pathway analyses were based on meta-analysis SNP p-values and all databases available in MAGENTA, including Gene Ontology, Ingenuity, KEGG and PANTHER. We set the threshold for significance at False Discovery Rate (FDR) < 0.05.
Second, we tested whether SNPs previously associated with OA in European Caucasian populations were also associated with radiographic knee OA in African Americans. We selected for replication 12 SNPs associated with knee OA at genome-wide levels of significance or near genome-wide significance in prior genome-wide association studies (GWAS) (8–12). In addition, we calculated the power to detect the association of each SNP with radiographic knee OA in our African American sample based on the odds ratio reported from the literature and the allele frequency of that SNP observed in African Americans (Table 4).
Table 4.
Association of knee OA in African Americans with SNPs robustly associated with knee OA in prior studies
| SNP effect in European Caucasians | SNP effect in African Americans | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr | Risk/other allele | Frequency of risk allele in ECs | OR | Ref | Frequency of risk allele in AAs | Power to detect OR of magnitude seen in ECs† | OR (95% CI)‡ |
P-value from meta-analysis |
| rs4730250* | 7q22 (DUS4L) | 7 | G/A | 0.17 | 1.17 | 9, 10 | 0.01 | 0.10 | NA | NA |
| rs143383 | GDF5 | 20 | T/C | 0.59–0.68 | 1.16 | 11 | 0.12 | 0.46 | 0.92 (0.68–1.24) |
0.59 |
| rs11842874 | MCF2L | 13 | A/G | 0.91–0.94 | 1.17 | 12 | 0.77 | 0.56 | 1.00 (0.80–1.26) |
0.98 |
| rs6976 | GLT8D1 | 3 | T/C | 0.37 | 1.12 | 8 | 0.15 | 0.34 | 0.99 (0.75–1.30) |
0.95 |
| rs11177 | GNL3 | 3 | A/G | 0.38 | 1.12 | 8 | 0.17 | 0.36 | 0.98 (0.75–1.27) |
0.87 |
| rs4836732 | ASTN2 | 9 | C/T | 0.47 | 1.20 | 8 | 0.67 | 0.79 | 1.11 (0.90–1.35) |
0.33 |
| rs9350591 | FILIP1; SENP6 | 6 | T/C | 0.11 | 1.18 | 8 | 0.12 | 0.54 | 0.90 (0.67–1.21) |
0.48 |
| rs10492367* | KLHDC5; PTHLH | 12 | T/G | 0.19 | 1.14 | 8 | 0.01 | 0.09 | NA | NA |
| rs835487 | CHST11 | 12 | G/A | 0.34 | 1.13 | 8 | 0.64 | 0.51 | 0.93 (0.77–1.15) |
0.53 |
| rs12107036 | TP63 | 3 | G/A | 0.52 | 1.21 | 8 | 0.35 | 0.90 | 1.12 (0.91–1.38) |
0.29 |
| rs8044769 | FTO | 16 | C/T | 0.50 | 1.11 | 8 | 0.76 | 0.32 | 0.94 (0.75–1.17) |
0.56 |
| rs10948172 | SUPT3H; CDC5L | 6 | C/T | 0.29 | 1.14 | 8 | 0.30 | 0.59 | 0.95 (0.77–1.16) |
0.60 |
Abbreviations: SNP = single nucleotide polymorphism; Chr = chromosome; EC = European Caucasians; OR = Odds ratio; Ref = reference; AA = African Americans; CI = confidence interval; NA = not applicable
SNPs with a minor allele frequency < 0.05 were excluded from the meta-analysis
Power based on 742 cases and 475 controls, p-value = 0.05, and population prevalence of knee OA = 0.35
Odds ratios adjusted for age, sex, and population stratification
Results
Sample characteristics of the 1,217 African American subjects included in this analysis (742 cases and 475 controls) are provided in Table 1. Compared to controls, cases were older, had higher BMI, and were more likely to be female. Approximately 50% of JoCo study subjects were cases compared to 72% of OAI study subjects. The larger number of cases than controls in OAI can be attributed to that study’s inclusion criteria, which were designed to enrich the cohort with subjects who had symptomatic radiographic knee OA.
Table 1.
Baseline characteristics of JoCo and OAI study participants
| JoCo | OAI | Combined | ||||
|---|---|---|---|---|---|---|
| Cases (n = 293) |
Controls (n = 297) |
Cases (n = 449) |
Controls (n = 178) |
Cases (n = 742) |
Controls (n = 475) |
|
| Age (yrs) | 64.0 (10.8) | 58.5 (9.6) | 59.6 (8.4) | 57.4 (8.3) | 61.4 (9.6) | 58.1 (9.1) |
| BMI (kg/m2) | 34.7 (8.8) | 30.1 (6.3) | 31.9 (4.6) | 29.0 (4.6) | 33.0 (6.7) | 29.7 (5.7) |
| % women | 68.9 | 61.9 | 69.7 | 61.2 | 69.4 | 61.7 |
Abbrevations: BMI = body mass index, JoCo = Johnston County Osteoarthritis Project, OAI = Osteoarthritis Initiative
We performed a genome-wide association analysis of radiographic knee OA to identify novel associations in African Americans. Based on the relatively small sample size, we regard these analyses as exploratory. The genomic inflation factor, λ, was 0.954 for the combined meta-analysis, providing no evidence for inflation of p-values. As shown in the meta-analysis GWAS results (Figure 1), one locus in LINC01006, a long intergenic non-protein coding RNA on chromosome 7, reached genome-wide significance in the primary meta-analysis (rs7792864, MAF=12%, OR [95% CI]=2.35 [1.77–3.13], p-value=4.11×10−9). Five other independent loci were associated with knee OA at p-values less than 1×10−6. These loci are located in or near MAGI1 (rs145965284, MAF=27%), ANKRD6 (rs78571182, MAF=12%), EPPK1/PLEC (rs76983122, MAF=11%), PAX7/TAS1R2 (rs4920343, MAF=13%), and DDX10/C11orf87 (rs9783397, MAF=29%); odds ratios ranged from 1.83 to 2.08 (Table 2). Regional association plots showed strong supporting signals in high linkage disequilibrium with rs7792864 (LINC01006) and rs76983122 (EPPK1/PLEC), but not rs78571182 (ANKRD6) (Figure 2). The reliability of the signal for rs145965284 (MAGI1) and rs9783397 (DDX10/C11orf87) was unclear due to lack of linkage disequilibrium data with other SNPs. After additional adjustment for BMI, odds ratios were slightly attenuated for associations in LINC01006, MAGI1, EPPK1/PLEC, and PAX7/TAS1R2, but not ANKRD6 or DDX10/C11orf87 (Table 2). P-values for heterogeneity ranged from 0.17 to 0.71, providing little evidence that there was significant heterogeneity between studies. In pathway analyses, we identified two pathways associated with knee OA that reached statistical significance (FDR < 0.05): 1) dorsal/ventral neural tube patterning and 2) iron ion transport (Table 3).
Figure 1.
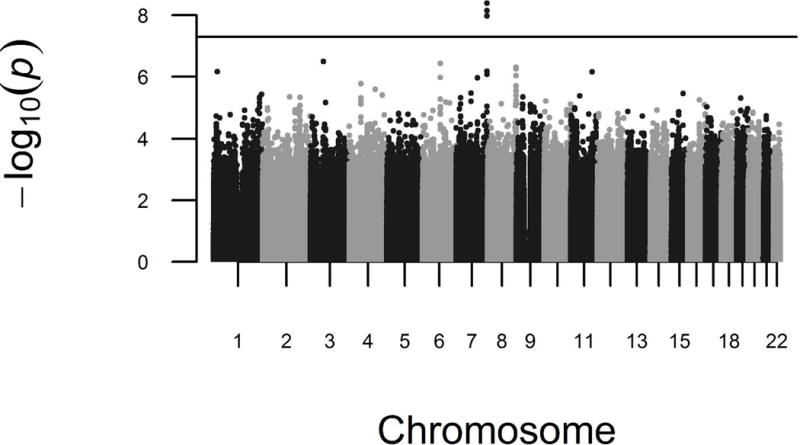
Manhattan plot of the knee OA GWAS meta-analysis in African Americans. The horizontal line represents the threshold for genome-wide significance, p-value < 5×10−8.
Table 2.
Associations with Knee OA in African Americans (p-value < 1×10−6)
| JoCo* | OAI* | Meta-Analysis, adjusted for age, sex, and PCs | Meta-Analysis, adjusted for age, sex, PCs, and BMI | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Nearest Gene(s) | Chr | Risk/Other Allele | Risk Allele Freq | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs7792864 | LINC01006 | 7 | C/G | 0.88 | 2.21 (1.43–3.41) |
3.56×10−4 | 2.47 (1.69–3.60) |
2.79×10−6 | 2.35 (1.77–3.13) |
4.11×10−9 | 2.29 (1.69–3.10) |
1.02×10−7 |
| rs145965284 | MAGI1 | 3 | A/T | 0.27 | 1.72 (1.22–2.44) |
1.94×10−3 | 2.27 (1.54−3.33) |
2.78×10−5 | 1.96 (1.51–2.54) |
3.18×10−7 | 1.92 (1.46–2.53) |
3.12×10−6 |
| rs78571182 | ANKRD6 | 6 | T/G | 0.88 | 2.59 (1.70–3.94) |
9.69×10−6 | 1.74 (1.19–2.54) |
4.25×10−3 | 2.08 (1.57–2.75) |
3.70×10−7 | 2.17 (1.61–2.93) |
3.62×10−7 |
| rs76983122 | EPPK1/PLEC | 8 | T/C | 0.11 | 2.70 (1.75–4.35) |
1.00×10−5 | 1.89 (1.18–3.03) |
4.25×10−3 | 2.31 (1.67–3.20) |
4.90×10−7 | 2.27 (1.60–3.21) |
3.76×10−6 |
| rs4920343 | PAX7/TAS1R2 | 1 | G/A | 0.87 | 1.79 (1.20–2.70) |
4.73×10−3 | 2.17 (1.52–3.13) |
3.43×10−5 | 2.00 (1.52–2.63) |
6.82×10−7 | 2.04 (1.54–2.78) |
1.27×10−6 |
| rs9783397 | DDX10/C11orf87 | 11 | T/G | 0.71 | 2.21 (1.53–3.19) |
4.73×10−3 | 1.59 (1.17–2.18) |
3.35×10−3 | 1.83 (1.44–2.32) |
6.99×10−7 | 1.88 (1.46–2.43) |
1.00×10−6 |
Abbreviations: Chr = chromosome, CI = confidence interval, Freq = frequency, JoCo = Johnston County Osteoarthritis Study, OAI = Osteoarthritis Initiative, OR = odds ratio, PCs = principal components
Odds ratios adjusted for age, sex, and population stratification
Figure 2.
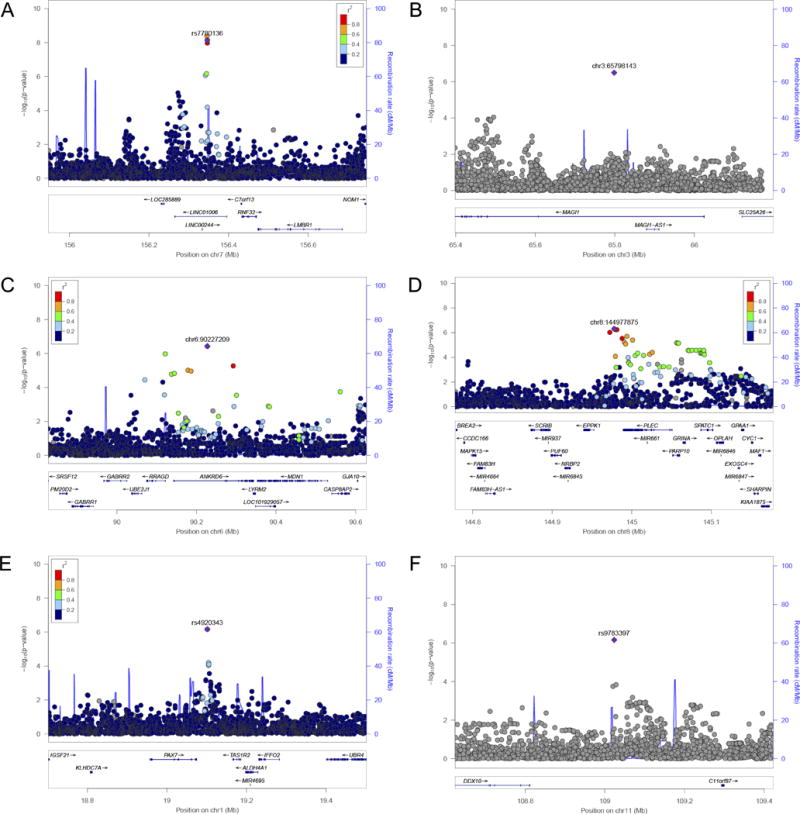
LocusZoom plots for top genetic associations with knee OA. –Log10 of the p-values are plotted against the genetic position (hg19) along the chromosome. Colored points represent the degree of linkage disequilibrium (r2) between the lead SNP (in purple) and all other SNPs in the region. Panels A-E are LocusZoom plots for LINC01006, MAGI1, ANKRD6, EPPK1/PLEC, PAX7/TAS1R2, and DDX10/C11orf87, respectively.
Table 3.
Pathways that Show Enrichment of Genes by MAGENTA Analysis
| Database | Gene Set | Effective Gene Set Size | No. of Expected Genes (>75% Cutoff) | No. of Observed Genes (>75% Cutoff) | Nominal P-value | FDR P-value | Significant Genes* |
|---|---|---|---|---|---|---|---|
| GOTERM | dorsal/ventral neural tube patterning | 13 | 3 | 11 | 1.00×10−5 | 5.60×10−3 | PAX7, SHH, GLI2, BMP4, PTCH1, SMO, MNX1, PSEN1, GSC, PSEN2, FOXA2 |
| GOTERM | iron ion transport | 22 | 6 | 15 | 1.00×10−4 | 1.09×10−2 | STEAP4, STEAP3, SLC25A28, SLC39A14, SLC40A1, SFXN5, FTL, TTYH1, STEAP2, MFI2, SFXN1, SFXN2, SFXN4, TF, FTH1 |
Enriched genes with scores in the top 25% of all gene scores. Best SNP p-values range from 1.68×10−3 to 1.27×10−6 for the dorsal/ventral neural tube patterning pathway and from 2.62×10−3 to 1.18×10−4 for the iron ion transport pathway.
Following the genome-wide association analysis, we attempted to replicate 12 SNPs previously identified to be robustly associated with OA, including 3 SNPs associated with knee OA (9–12) and 9 genome-wide or near genome-wide significant SNPs identified by the arcOGEN (Arthritis Research Council Osteoarthritis Genetics) study, the largest GWAS of OA in European Caucasians to date (8). Risk allele frequencies were lower in African Americans than European Caucasians for most SNPs (chromosome 7q22, GDF5, MCF2L, GLT8D1, GNL3, KLHDC5/PTHLH, and TP63). We did not test SNPs in chromosome 7q22 or KLHDC5/PTHLH due to minor allele frequencies less than 1%. Of the remaining 10 SNPs, none was significantly associated with knee OA in African Americans (Table 4). Power to detect significant associations of the same magnitude as seen in European Caucasians at p-value < 0.05 ranged from 32% to 90%. Power to detect nominal associations was greater than 80% for only one locus, TP63.
Discussion
This GWAS meta-analysis is the first genome-wide effort to identify genetic polymorphisms associated with knee OA in African Americans. We identified one genome-wide significant locus in LINC01006 and five loci that reached suggestive significance.
Pathway analyses suggested that dorsal/ventral neural tube patterning and iron ion transport pathways may be associated with knee OA in African Americans. However, we found little evidence that previously reported OA susceptibility variants in European Caucasians were associated with knee OA in African Americans. This suggests that the genetic architecture of knee OA in African Americans may be different from European Caucasians, warranting future genetic studies of knee OA in populations with African ancestry.
We identified a novel genome-wide significant locus in LINC01006, a long intergenic non-protein coding RNA located on chromosome 7q36 near INSIG2, SHH, C70rf13, RNF32, and LMBR1. The function of LINC01006 is not entirely clear, but it is increasingly appreciated that long intergenic non-protein coding RNAs may play key regulatory roles in gene expression, particularly among nearby genes (17). The locus on chromosome 7q36 has been linked to preaxial polydactyly and other limb disorders, highlighting the possibility that genes and regulatory elements found in this region are related to limb development (18–20). The chromosome 7q36 locus has also been implicated in genetic linkage studies of BMI (21, 22). In this GWAS, adjustment for BMI attenuated the association between LINC01006 variants and knee OA, suggesting that this locus could act in part through body weight regulation. Since overweight and obesity are highly prevalent in African Americans and also strong risk factors for knee OA (23, 24), it is possible that some knee OA loci discovered in African Americans may operate through their effects on BMI. After adjusting for BMI, most top loci had attenuated effects and no locus reached genome-wide significance.
None of the candidate SNPs that have been robustly associated with OA in prior studies of European Caucasian populations was replicated in this study of African Americans. The most likely reason for lack of replication is the relatively low power of our sample – even comprising 742 carefully phenotyped cases – to detect SNPs having relatively modest effect sizes (i.e., odds ratios ranging from 1.11 to 1.21). While the small sample size of our study limited ability to detect significant p-values, effect size that is independent from sample size may still be a good representation of the magnitude of effect on knee OA. Notably, the two loci for which power was sufficient for detecting an association (79% power for the risk allele in ASTN2 and 90% power for the risk allele in TP63), both had effect sizes consistent with that found in European Caucasians (odds ratios of 1.11 and 1.12, respectively). The odds ratios for other candidate loci, including GDF5, MCF2L, GLT8D1, GNL3, FILIP1/SENP6, CHST11, FTO, and SUPT3H/CDC5L were largely null and inconsistent with effect sizes identified in European Caucasians. The lack of robust replication could be a consequence of low power and/or heterogeneity in the JoCo and OAI populations, or indicate a difference in the genetic architecture of OA between African Americans and European Caucasians.
There may be differences in the underlying linkage disequilibrium structure between African and European populations that may alter the degree to which the tested SNPs tag the unmeasured pathogenic variant that is responsible for OA. Risk allele frequencies for 10 of 12 OA candidate SNPs were noticeably different between ethnic groups, including 7 SNPs that had lower frequencies and 3 SNPs that had higher frequencies in African Americans compared to Caucasians. The genome-wide significant variant that we identified in LINC01006 had a minor allele frequency of 12% in African Americans and was much less common in European Caucasian populations, where the minor allele frequency was less than 3%. Similarly, allele frequencies for other top findings were noticeably different between African Americans and European Caucasians. None of the top African American GWAS loci was associated with knee OA in our previous report of genetic associations in Caucasians (25).
It is also possible that the etiology of OA may be slightly different across ethnic groups. For example, African Americans experience a higher prevalence of knee OA (13), greater lateral tibiofemoral joint space narrowing, greater valgus thrust, and more pain than Caucasian Americans, which may be due to either biological or sociocultural differences, or both. However, African-Americans are less likely than Caucasian Americans to undergo total knee arthroplasty (14).
We had limited power to detect genome-wide significant loci given the small sample size, requiring odds ratios ranging from 1.51–1.65 in order to obtain significant associations in the GWAS; this is a much larger effect size than reported for all prior detected associations in European Caucasians. Effect sizes for all genome-wide significant and suggestive loci in this study were larger than 1.83, exceeding the minimum effect size needed to claim statistical significance with 80% power. This demonstrates an inherent bias in our study. We are more likely to identify variants with large effect sizes than modest effect sizes. It is possible that our top findings may actually have modest effects and by chance have detectable large effects in JoCo and OAI. Because of this, future replication of these associations in other cohorts will be essential. This phenomenon of the “winner’s curse” occurs often in GWAS and underscores the need to include even larger studies of knee OA in African Americans to replicate and refine genetic associations.
A strength of our study was the standardized and careful phenotyping of radiographic knee OA used in both JoCo and OAI. However, such assessments are costly and time-consuming to obtain, making it difficult to accrue the very large sample sizes needed for genetic association studies. Despite these difficulties, there is great scientific value to including non-European populations in genetic studies because associations with novel loci not previously detected in Europeans can sometimes be uncovered due to differences in allele frequencies between European and non-European populations or to differences in linkage disequilibrium structure. Association testing at established loci in populations representing different ethnic groups may also be useful in efforts to identify the causative variant. Even larger studies of OA genetics in African Americans are needed and have the potential to provide insight into disparities in OA prevalence and etiology. We invite other researchers to contribute additional samples to a larger scale meta-analysis of OA in under-represented populations.
In summary, we conducted an exploratory GWAS meta-analysis study of radiographic knee OA in African Americans in two of the largest, well-characterized OA cohorts. We identified a genome-wide significant finding in LINC01006 and significant pathways in dorsal/ventral neural tube patterning and iron ion transport, but did not replicate associations with SNPs previously associated with OA in European populations possibly due to lack of power. These results highlight the importance of including non-European ancestry populations in studies of OA genetics.
Acknowledgments
Funding: The Genetic Components of Knee Osteoarthritis (GeCKO) project was supported by the American Recovery and Reinvestment Act (ARRA) through grant number RC2-AR-058950 from NIAMS/NIH. The OAI is public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the NIH. The JoCo is supported in part by S043, S1734, & S3486 from the CDC/Association of Schools of Public Health; 5-P60-AR30701 & 5-P60-AR49465-03 from NIAMS/NIH and Algynomics, Inc. Partial analysis support was provided by P30 DK072488. MSY was supported by NIA/NIH T32AG00262, Arthritis Foundation Doctoral Dissertation Award (6081), NIA/NIH T32AG023480, and Friends of Hebrew SeniorLife.
Footnotes
Contributions
Study concept and design: MCH, BDM, RDJ, and JMJ. Data collection: DJD, JBR, MCH, BDM, RDJ, and JMJ. Analysis and interpretation: YL, MSY, LMY-A, DJD, JBR, MCH, BDM, RDJ, and JMJ. Drafting manuscript: YL, MSY, MCH, BDM, RDJ, and JMJ. Critical review and final approval of manuscript content: YL, MSY, LMY-A, DJD, JBR, MCH, BDM, RDJ, and JMJ. YL, MSY, and JMJ take responsibility for the data and accuracy of the data analyses.
Competing interests
All authors report no relevant competing interests.
Disclosures: None of the authors has any relevant conflicts of interest to report.
References
- 1.Hirsch R, Lethbridge-Cejku M, Hanson R, Scott WW, Reichle R, Plato CC, et al. Familial aggregation of osteoarthritis: data from the Baltimore Longitudinal Study on Aging. Arthritis Rheum. 1998;41:1227–32. doi: 10.1002/1529-0131(199807)41:7<1227::AID-ART13>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 2.Spector TD, Cicuttini F, Baker J, Loughlin J, Hart D. Genetic influences on osteoarthritis in women: a twin study. BMJ. 1996;312:940–3. doi: 10.1136/bmj.312.7036.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manek NJ, Hart D, Spector TD, MacGregor AJ. The association of body mass index and osteoarthritis of the knee joint: an examination of genetic and environmental influences. Arthritis Rheum. 2003;48:1024–9. doi: 10.1002/art.10884. [DOI] [PubMed] [Google Scholar]
- 4.Styrkarsdottir U, Thorleifsson G, Helgadottir HT, Bomer N, Metrustry S, Bierma-Zeinstra S, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet. 2014;46:498–502. doi: 10.1038/ng.2957. [DOI] [PubMed] [Google Scholar]
- 5.Castano Betancourt MC, Cailotto F, Kerkhof HJ, Cornelis FM, Doherty SA, Hart DJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci U S A. 2012;109:8218–23. doi: 10.1073/pnas.1119899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evangelou E, Valdes AM, Castano-Betancourt MC, Doherty M, Doherty S, Esko T, et al. The DOT1L rs12982744 polymorphism is associated with osteoarthritis of the hip with genome-wide statistical significance in males. Ann Rheum Dis. 2013;72:1264–5. doi: 10.1136/annrheumdis-2012-203182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evangelou E, Kerkhof HJ, Styrkarsdottir U, Ntzani EE, Bos SD, Esko T, et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis. 2014;73:2130–6. doi: 10.1136/annrheumdis-2012-203114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.arcOGEN Consortium. Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–23. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, Arp P, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010;62:499–510. doi: 10.1002/art.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evangelou E, Valdes AM, Kerkhof HJ, Styrkarsdottir U, Zhu Y, Meulenbelt I, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis. 2011;70:349–55. doi: 10.1136/ard.2010.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdes AM, Evangelou E, Kerkhof HJ, Tamm A, Doherty SA, Kisand K, et al. The GDF5 rs143383 polymorphism is associated with osteoarthritis of the knee with genome-wide statistical significance. Ann Rheum Dis. 2011;70:873–5. doi: 10.1136/ard.2010.134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day-Williams AG, Southam L, Panoutsopoulou K, Rayner NW, Esko T, Estrada K, et al. A variant in MCF2L is associated with osteoarthritis. Am J Hum Genet. 2011;89:446–50. doi: 10.1016/j.ajhg.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan JM, Helmick CG, Renner JB, Luta G, Dragomir AD, Woodard J, et al. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project. J Rheumatol. 2007;34:172–80. [PubMed] [Google Scholar]
- 14.Centers for Disease C, Prevention. Racial disparities in total knee replacement among Medicare enrollees–United States, 2000–2006. MMWR Morb Mortal Wkly Rep. 2009;58:133–8. [PubMed] [Google Scholar]
- 15.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 18.Heutink P, Zguricas J, van Oosterhout L, Breedveld GJ, Testers L, Sandkuijl LA, et al. The gene for triphalangeal thumb maps to the subtelomeric region of chromosome 7q. Nat Genet. 1994;6:287–92. doi: 10.1038/ng0394-287. [DOI] [PubMed] [Google Scholar]
- 19.Hing AV, Helms C, Slaugh R, Burgess A, Wang JC, Herman T, et al. Linkage of preaxial polydactyly type 2 to 7q36. Am J Med Genet. 1995;58:128–35. doi: 10.1002/ajmg.1320580208. [DOI] [PubMed] [Google Scholar]
- 20.Zguricas J, Heus H, Morales-Peralta E, Breedveld G, Kuyt B, Mumcu EF, et al. Clinical and genetic studies on 12 preaxial polydactyly families and refinement of the localisation of the gene responsible to a 1.9 cM region on chromosome 7q36. J Med Genet. 1999;36:32–40. [PMC free article] [PubMed] [Google Scholar]
- 21.Kettunen J, Perola M, Martin NG, Cornes BK, Wilson SG, Montgomery GW, et al. Multicenter dizygotic twin cohort study confirms two linkage susceptibility loci for body mass index at 3q29 and 7q36 and identifies three further potential novel loci. Int J Obes (Lond) 2009;33:1235–42. doi: 10.1038/ijo.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DF, Pang Z, Li S, Thomassen M, Wang S, Jiang W, et al. High-resolution genome-wide linkage mapping identifies susceptibility loci for BMI in the Chinese population. Obesity (Silver Spring) 2012;20:830–3. doi: 10.1038/oby.2010.349. [DOI] [PubMed] [Google Scholar]
- 23.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA. 2014;311:806–14. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hootman JM, Helmick CG, Hannan CJ, Pan LP. Prevalence of Obesity Among Adults With Arthritis-United States, 2003–2009 (Reprinted from MMWR, vol 60, pg 509–513, 2011) JAMA. 2011;305:2404–5. [PubMed] [Google Scholar]
- 25.Yau MS, Yerges-Armstrong LM, Liu Y, Lewis CE, Duggan DJ, Renner JB, et al. Genome-Wide Association Study of Radiographic Knee Osteoarthritis in North American Caucasians. Arthritis Rheumatol. 2017;69:343–51. doi: 10.1002/art.39932. [DOI] [PMC free article] [PubMed] [Google Scholar]


