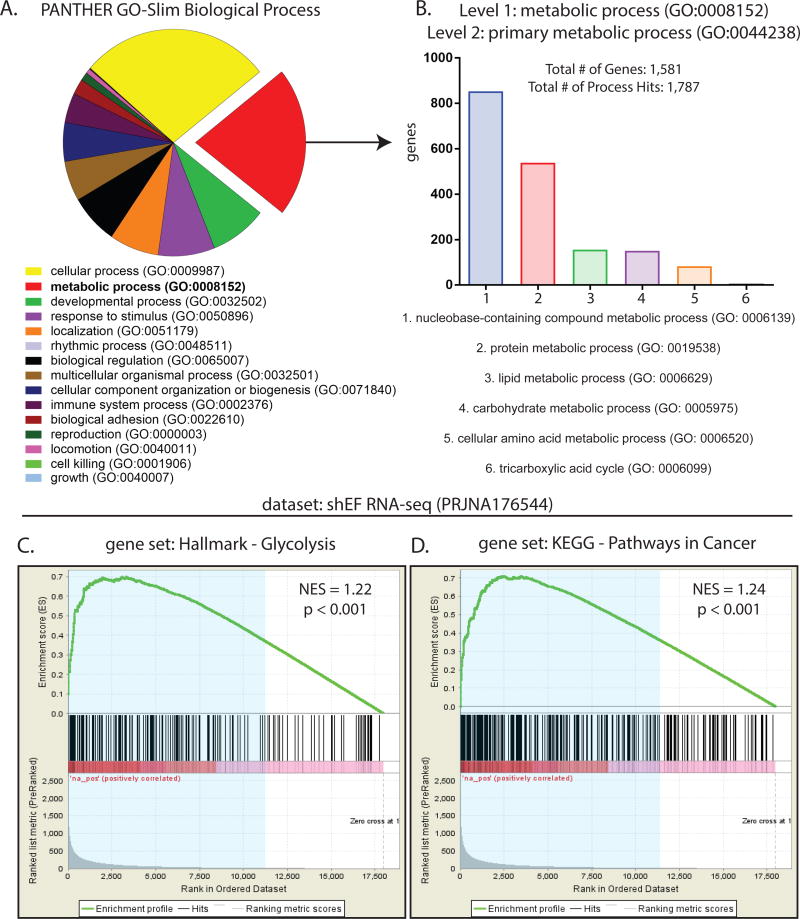
Figure 1. EWS/FLI exerts significant effects on expression of genes important for metabolism.
A. Gene ontology analysis of 5,698 EWS/FLI-regulated genes from publicly available RNA-seq data (27,37) using the PANTHER GO-Slim Biological Process annotation dataset. Threshold of significance for differential expression was log2(ratio) ≥ 0.585 and AdjP ≥ 10. Of these, 1,920 (33.7%) were annotated as metabolic processes (GO: 0008152). B. Among these metabolic processes (GO: 0008152), genes annotated as primary metabolic processes (GO: 0044238) were mostly involved in nucleobase-containing compound metabolism (GO: 0006139) and protein metabolism (GO: 0019538). C & D. Gene set enrichment analysis comparing a rank-ordered dataset (ranked by AdjP) of EWS/FLI knockdown RNA-seq data to the gene sets Hallmark-Glycolysis and KEGG-Pathways In Cancer, available from the Molecular Signatures Database (MSigDB) version 5.2. Differentially expressed genes (blue shaded region) in shEF vs. shLUC correlate significantly with genes important for glycolysis and pathways commonly altered in cancer. NES = normalized enrichment score.