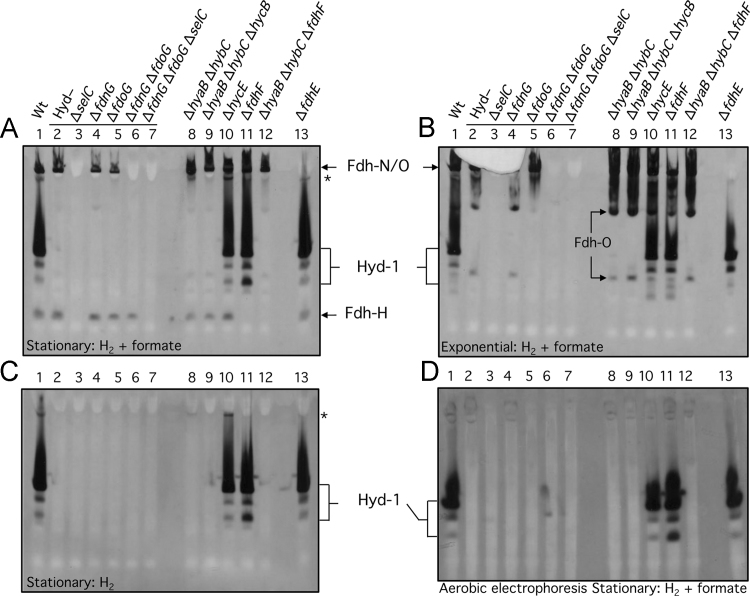
Fig. 2.
Identification of conditions revealing the activity of the Fdh-H enzyme. Crude extracts (30 µg of protein) derived from the anaerobically grown E. coli strains indicated were subjected to 6% (w/v polyacrylamide) non-denaturing PAGE and subsequently stained for Fdh and Hyd enzyme activities (A, B, D) or only Hyd activity (C) using the PMS-NBT staining procedure (see Materials and methods). The gels shown in panels A, B, and C were run anaerobically, while that shown in panel D was run aerobically. Cells were harvested when the cultures reached the exponential phase (B) or the stationary phase (A, C, D) of growth. The phase of growth and the electron donors used for staining are indicated at the bottom of each panel. The strains included: Lane 1, MC4100 (wild type); lane 2, FTD147 (ΔhyaB, ΔhybC, ΔhycE); lane 3, SH1 (ΔhyaB, ΔhybC, ΔhycE, ΔselC); lane 4, SH173 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG); lane 5, SH174, (ΔhyaB, ΔhybC, ΔhycE, ΔfdoG); lane 6, SH175 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG, ΔfdoG); lane 7, SH200 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG, ΔfdoG, ΔselC); lane 8, CP734 (ΔhyaB, ΔhybC); lane 9, CP1002 (ΔhyaB, ΔhybC, ΔhycB); lane 10, HD705 (ΔhycE); lane 11, CP585 (ΔfdhF); lane 12, CP1010 (ΔhyaB, ΔhybC, ΔfdhF); lane 13, JW3862 (ΔfdhE). The phenotype or genotype of the respective strain is indicated above the lanes. Note that the horizontal line above lanes 2–7 in A and B indicates that all strains lacked active Hyd-1, -2, and -3. The activity band(s) due to the respective enzyme complexes are indicated beside the panels and the asterisk denotes an unidentified H2-dependent enzyme complex. The migration of two faster-migrating Fdh-O-dependent enzyme complexes observed in exponentially growing cells (B) is indicated within the figure.