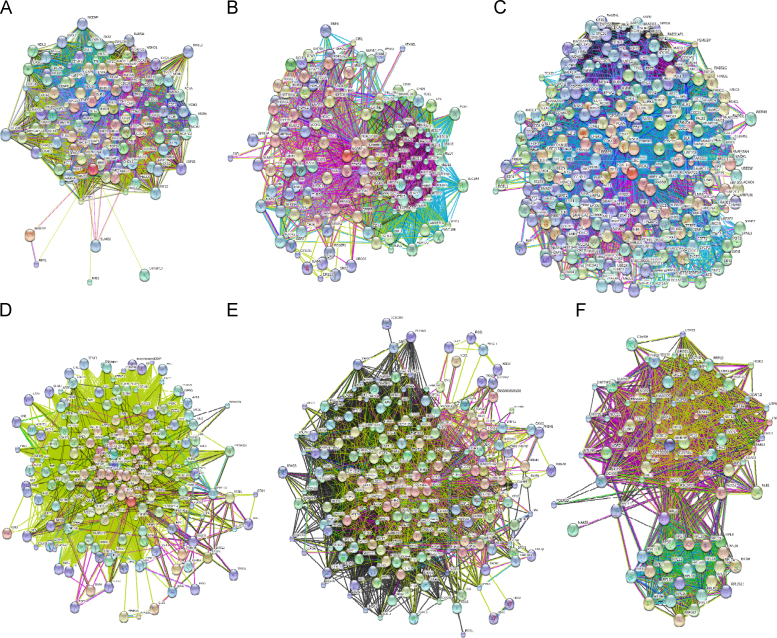
Fig. 6.
Analysis of the interactivity of the several human nuclear proteins by the STRING platform [58]. Analyzed proteins are: (A) KI-67 (UniProt ID: P46013), (B) N-CoR2 (UniProt ID: Q9Y618), (C) BRCA1 (UniProt ID: P38398), (D). SON (UniProt ID: P18583), (E) ATRX (UniProt ID: P46100), and (F) RLP24 (UniProt ID: Q9UHA3). STRING produces the network of predicted associations for a particular group of proteins. The network nodes are proteins, whereas the edges represent the predicted or known functional associations. An edge is drawn with up to 7 differently colored lines that represent the existence of the seven types of evidence used in predicting the associations. A red line indicates the presence of fusion evidence; a green line – neighborhood evidence; a blue line – co-occurrence evidence; a purple line – experimental evidence; a yellow line – text mining evidence; a light blue line – database evidence; a black line – co-expression evidence [58].