Figure 5.
Translation Alterations in SMA Reveal Links between SMN and Ribosome Biology
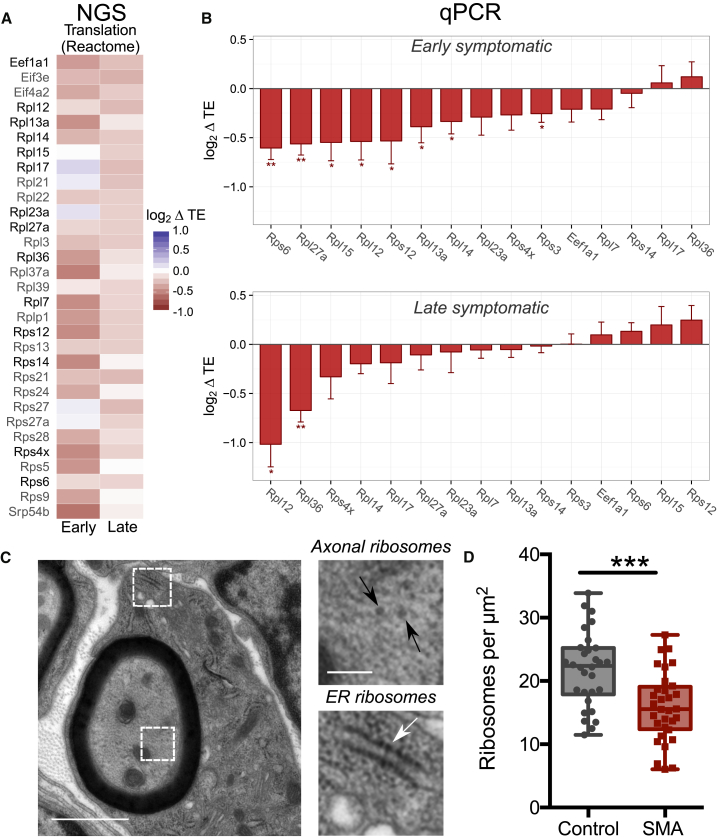
(A) Heatmap displaying all genes with altered translation efficiency (ΔTE) in SMA brains and annotated under the “Translation” Reactome pathway, significantly enriched in Figure 4D. The majority of genes belong to the family of ribosomal proteins. Genes further analyzed by qPCR are highlighted in black.
(B) qPCR-derived variations of translation efficiency for ribosomal proteins and one elongation factor from (A) in early (top) and late (bottom) symptomatic mouse brains. Mean value ± SEM is shown; three to four biological replicates and two to six technical replicates; all genes were normalized to the geometric mean of actin and cyclophilin a; one-tailed t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Representative electron micrograph of a large diameter (motor) axon in the intercostal nerve from a P5 SMA mouse (black arrowheads: axonal ribosomes; white arrowhead, ER ribosomes).
(D) Counts of axonal ribosomes revealed a decrease in the density of ribosomes in SMA mouse axons compared to controls (n = 30 axon profiles; N = 3 mice per genotype; ∗∗∗p < 0.001, two-tailed t test).
See also Figure S5.