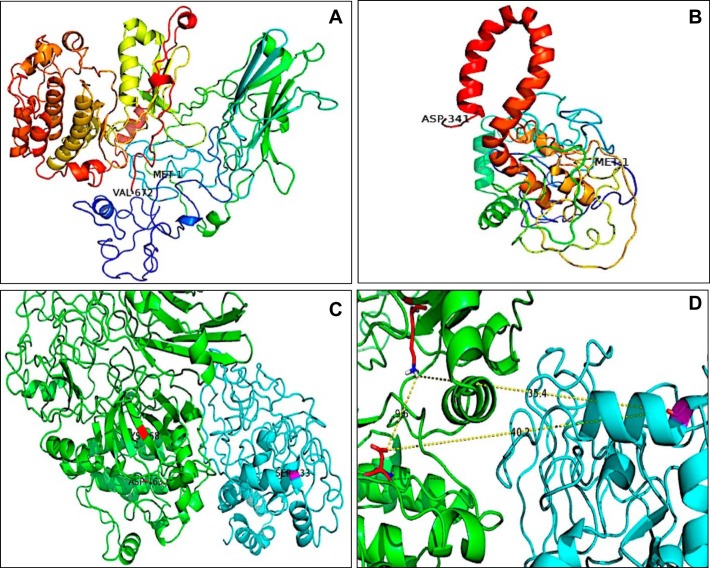
Fig. 8.
Three-dimensional in silico protein–protein docking of PKCα and CREB1. A and B: the 3-dimensional structures were built using the web server Phyre2. N-Terminal and C-terminal amino acids are shown in the images and correspond to MET 1 and VAL 672 for PKCα and MET 1 and ASP 341 for CREB1. C: analysis performed with ClusPro software. PKCα is shown in green, and CREB1 is shown in cyan. D: amino acids involved in the catalytic reaction in PKCα: LYS 368 and ASP 463, in close proximity with Ser133 of CREB1 (35.4 and 40.2 Å, respectively).