Abstract
Antibodies have widespread applications in areas ranging from therapeutics to chromatography and protein microarrays. Certain applications require only the fragment antigen-binding (Fab) units of the protein. This study compares the cleavage efficacy of dithiothreitol (DTT), mercaptoethylamine (MEA), and dithiobutylamine (DTBA) – a relatively new reducing agent synthesized in 2012. Pseudo-first order kinetic analyses show DTBA to be ~213 times faster than DTT and ~71 times faster than MEA in the formation of Fab׳ antibody fragments from polyclonal rabbit antibodies. Monoclonal mouse antibodies were also used to show the feasibility of the reduction process on antibodies from a different species and with a different clonality. DTBA cleaved the monoclonal mouse F(ab)2 units most efficiently, ~2 times faster than DTT ~10 times faster than MEA. Due to the extremely quick reactivity of all the reducing agents in the first five minutes of monoclonal antibody reductions as well as for the DTBA reductions of the polyclonal rabbit antibodies, the pseudo-first order kinetic analyses should be interpreted qualitatively for these results. Nucleophilic sulfides on Fab׳ fragments are preserved in the DTBA reduction process, demonstrated by their reactivity with Ellman׳s reagent. Degradation of the Fab׳ fragments was observed with the monoclonal mouse antibodies after reduction with DTBA or DTT. In conclusion, DTBA is the more efficient reducing agent compared to DTT and MEA, however, the reduction process should be optimized as degradation of the Fab׳ fragments is possible.
Keywords: Fab׳ fragments, Antibody reduction, Dithiobutylamine, Dithiothreitol, Mercaptoethylamine
Graphical abstract
Antibodies have widespread applications in areas ranging from therapeutics to chromatography and protein microarrays. Certain applications require only the fragment antigen-binding (Fab) units of the protein. This study compares the cleavage efficacy of dithiothreitol (DTT), mercaptoethylamine (MEA), and dithiobutylamine (DTBA) – a relatively new reducing agent synthesized in 2012. Pseudo-first order kinetic analyses show DTBA to be ~213 times faster than DTT and ~71 times faster than MEA in the formation of Fab׳ antibody fragments from polyclonal rabbit antibodies. Monoclonal mouse antibodies were also used to show the feasibility of the reduction process on antibodies from a different species and with a different clonality. DTBA cleaved the monoclonal mouse F(ab)2 units most efficiently, ~2 times faster than DTT ~10 times faster than MEA. Due to the extremely quick reactivity of all the reducing agents in the first five minutes of monoclonal antibody reductions as well as for the DTBA reductions of the polyclonal rabbit antibodies, the pseudo-first order kinetic analyses should be interpreted qualitatively for these results. Nucleophilic sulfides on Fab׳ fragments are preserved in the DTBA reduction process, demonstrated by their reactivity with Ellman׳s reagent. Degradation of the Fab׳ fragments was observed with the monoclonal mouse antibodies after reduction with DTBA or DTT. In conclusion, DTBA is the more efficient reducing agent compared to DTT and MEA, however, the reduction process should be optimized as degradation of the Fab׳ fragments is possible.
Highlights
-
•
Dithiobutylamine (DTBA) is a relatively new reducing agent synthesized in 2012.
-
•
Antibody cleavage efficiency was compared with DTT, MEA, and DTBA.
-
•
DTBA was able to cleave monoclonal mouse and polyclonal rabbit antibodies.
-
•
Fab׳ nucleophilic sulfides were preserved during the cleavage process.
-
•
DTBA cleavage should be optimized as undesirable byproducts are possible.
1. Introduction
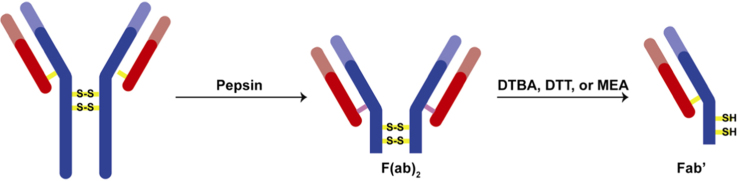
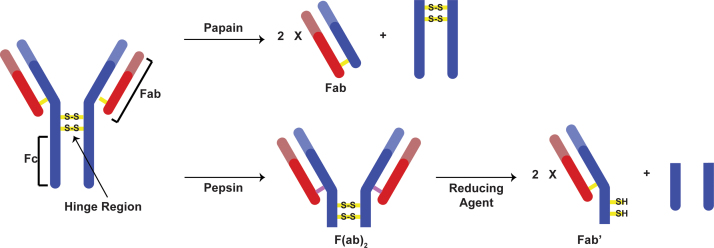
Immunoglobulins, or antibodies, are large proteins produced by the immune system that have found numerous applications in many different fields. Some examples include the use of these proteins in therapeutics, immunoaffinity capillary electrophoresis, immunoaffinity chromatography, protein microarray technology, and immunosensors [1], [2], [3], [4], [5]. Certain applications require the use of only specific parts of the antibody, such as the F(ab)2, Fab, or Fc fragments (Fig. 1) [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16].
Fig. 1.
Cleavage scheme for a general IgG antibody using either papain or pepsin and a reducing agent.
One of the most commonly encountered classes of antibodies is the immunoglobulin G (IgG). The structure of a general IgG antibody contains four protein chains – two pairs of two chains, with each pair consisting of a heavy and light chain [17], [18]. The portion of the antibody that binds to specific antigens is called the fragment–antigen binding (Fab) unit. Each antibody contains two Fab fragments and one tail Fc fragment (Fig. 1). The proline and cysteine-rich hinge region contains key disulfide bridges that hold the two halves of the antibody together [17], [18]. This region lies exposed between the heavy chain constant variable domains CH1 and CH2, making it susceptible for enzymatic attack [17].
A common approach for the fragmentation of antibodies is through the use of proteolytic enzymes [19], [20], [21], [22], [23]. Papain, pepsin, bromelain, ficin, lysyl endopeptidase are frequently used enzymes, with the first two being the most popular. Antibody cleavage with papain results in the formation of two Fab fragments and one Fc fragment [19], [22], [23]. This enzyme cleaves the antibody above the two key hinge disulfide bridges (Fig. 1). Alternatively, pepsin cleaves the antibody below the two disulfide bridges, resulting in an F(ab)2 fragment and an Fc fragment [20], [21], [22], [23]. The F(ab)2 fragment may then be reduced to yield two Fab׳ fragments with C-terminal nucleophilic sulfides [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]. Pepsin may also be immobilized onto a resin instead of dissolved in solution [2]. This allows for the reaction to be stopped immediately through the quick removal of the resin from solution. However, antibody cleavage using either papain or pepsin must be optimized for each antibody class and subtype [21], [22], [23]. For example, mouse IgG1 cannot be cleaved using pepsin whereas mouse IgG2a and IgG2b are very reactive towards pepsin [21]. Although pepsin has an optimum cleavage efficiency in very acidic solutions (pH=2.0), antibody denaturation and diminished antigen-binding ability are also associated with such harsh conditions [23]. As a result, less acidic (pH=3–4) solutions are used during pepsin antibody cleavage.
The reduction of F(ab)2 dimers, formed from the pepsin cleavage, yields Fab׳ fragments containing nucleophilic sulfides that can be used successfully for immobilization onto surfaces [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]. Some commonly encountered reducing agents are β-mercaptoethanol (β-ME), tris(2-carboxyethyl)phosphine (TCEP), dithiothreitol (DTT), and mercaptoethylamine (MEA) [24], [25], [26], [27], [28], [29]. The formation of Fab׳ fragments from F(ab)2 involves the initial rate-limiting nucleophilic attack of the reducing agent on a hinge disulfide bond [27]. After the steps for thiol-disulfide interchange are completed, Fab׳ fragments are released. Monothiol reducing agents, such as β-ME, have been shown to “adhere” to the molecules that they are reducing and are not preferred for F(ab)2 cleavage [24]. Phosphine-based reducing agents, such as TCEP, offer the benefit of yielding irreversible reactions, driven by the formation of trialkylphosphine oxides. TCEP is also air-stable, reacts rapidly, and can be used in dilute conditions [26]. Although more efficient than DTT at cleaving small molecule disulfides, TCEP has significant limitations with large peptides due to steric crowding from the 2-carboxyethyl arms [24], [29]. Currently, the two most common reducing agents for the formation of Fab׳ fragments are DTT and MEA. As a dithiol, DTT offers the benefit of performing an intramolecular attack of the disulfide once the first thiol has attached to the molecule being reduced. The resulting oxidized DTT product is a cyclic disulfide that is thermodynamically favorable due to its steric specifications [25]. This intramolecular step is roughly 103–104 faster compared to its intermolecular counterpart [27]. Another benefit of DTT is that it is possible to monitor the course of the reaction by UV–vis methods through the formation of the oxidized DTT molecules, which absorb at 310 nm. MEA, although a monothiol, is able to cleave F(ab)2 dimers without “sticking” to the molecules [1], [2]. Compared to MEA, DTT has shown to be inconsistent with the cleavage of different antibodies, sometimes cleaving the disulfide bonds between the antibody chains [2].
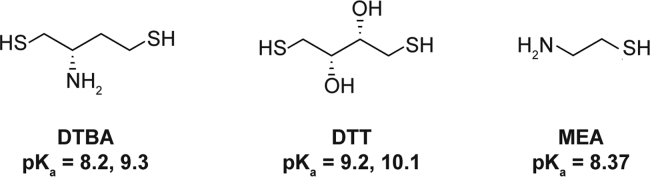
First synthesized in 2012, dithiobutylamine (DTBA) is a new dithiol reducing agent, similar in structure to both DTT and MEA [24], [30]. Due to its novelty, it has had limited application and has not been used yet, to our knowledge, in the field of antibody-based chemistry. However, DTBA has desirable characteristics since its hydrochloride salt is nearly odorless and is highly soluble in water [24]. Having lower pKa values allows for DTBA to have more nucleophilic deprotonated sulfides present at any given pH compared to DTT and MEA (Fig. 2). The lower pKa values are believed to be a result of strong Coulombic and inductive effects from the amino group, similar to what is observed with MEA [24]. The amino group of DTBA may also be used to immobilize DTBA to resins. Immobilized DTBA has been used in reducing cystines of large enzymes, although it has diminished cleaving ability compared to DTBA molecules free in solution [30].
Fig. 2.
Structures and pKas of DTBA, DTT, and MEA [24].
In this study, the reduction efficacies of DTT, MEA, and DTBA are kinetically compared with respects to the cleavage of polyclonal rabbit IgG F(ab)2 fragments. The three reducing agents are compared at room temperature (22 °C) and physiological temperature (37 °C). Reductions with DTBA are also compared using varying concentrations of the reducing agent at room temperature. Reduced solutions are analyzed quantitatively using size-exclusion chromatography and UV–vis spectroscopy and qualitatively with sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE). The Fab׳ fragments are reacted with Ellman׳s reagent to determine the preservation of the C-terminal nucleophilic sulfides [27], [31], [32]. In order to test the feasibility of cleaving antibodies from other species and with differing clonalities, monoclonal mouse anti-human IgG1 antibodies were also examined. By virtue of developing reaction conditions for the kinetic study of the three reducing agents, a new protocol for cleaving antibodies using DTBA was created.
2. Materials and methods
2.1. Materials
Polyclonal rabbit anti-goat IgG antibodies (G5518) and monoclonal mouse anti-human IgG1 antibodies (I2513) were purchased from Sigma and used without further purification. Pepsin from porcine gastric mucosa, MEA, DTT, and DTBA were also purchased from Sigma. Slide-A-Lyzer dialysis cassettes (2000 Da MWCO) were obtained from Thermo Scientific. BenchMark Protein Ladder was received from Life Technologies. Acrylamide, bisacrylamide, glycine, sodium dodecyl sulfate (SDS), methanol, glacial acetic acid, Coomassie blue, bromophenol blue, glycerol, iodoacetamide, 5–5׳-dithiobis(2-nitrobenzoic acid) (Ellman׳s reagent), sodium acetate, Trizma base, ethylenediaminetetraacetic acid (EDTA), sodium chloride, sodium citrate tribasic dehydrate, sodium phosphate dibasic, and sodium tetraborate decahydrate were purchased from Sigma and used without further purification. Deionized water, 18.2 Ω, was obtained from a Milli-Q integral water purification system. UV–vis analyses were performed on an Agilent Cary 60 UV–vis spectrophotometer operating with a xenon lamp. HPLC purification was performed at St. Michael׳s Hospital (Toronto, Canada), using two in-line Agilent ZORBAX Bio Series GF-250 size exclusion columns and a UV–vis detector. Columns measured 250 mm in length, with an internal diameter of 4.6 mm. The columns were packed with 4 µm spherical silica having a pore size of 150 Å. The separations were performed at room temperature with 0.5 mL/min flow rates and 100 µL injection volumes using PBS (100 mM sodium phosphate dibasic, pH 7.0) mobile phase. The peaks were monitored at 280 nm wavelengths using yeast (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome C (12 kDa) calibration standards (2.5 mg/mL each).
2.2. SDS-PAGE analyses
All samples for SDS-PAGE were prepared in non-reducing conditions. The analyses were cast using 12% SDS-PAGE gels from a 40% acrylamide solution (36:1 acrylamide:bisacrylamide mass ratio). Samples were run at constant voltage (220 V) using Laemmli running buffer. The non-reducing loading buffer, containing 11.5% (v/v) 1.0 M Trizma pH 8.8 buffer, 3.7% (w/v) SDS, 27.5% (v/v) glycerol, and 0.018% (w/v) bromophenol blue, was prepared in distilled water. The Benchmark Protein Ladder (10–190 kDa) was used for molecular weight sizing. A 0.05% (w/v) Coomassie blue stain containing 40% (v/v) methanol and 10% (v/v) glacial acetic acid was used to stain the gels. To destain the gels, a solution containing 40% (v/v) methanol and 10% (v/v) glacial acetic acid in distilled water was used.
2.3. Optimization of whole antibody to F(ab)2 cleavage
Enzymatic digestion of polyclonal anti-goat IgG and monoclonal anti-human IgG1 using pepsin was tested with both phosphate-buffered saline (PBS) (10 mM sodium phosphate dibasic, 154 mM sodium chloride) and acetate (100 mM sodium acetate) buffers at pH 4.0 and 4.5. The following incubation incubation times were tested for each buffer and pH combination: 1, 2, 3, 4, 6, 8, 24, and 48 h. A 1 mg/mL antibody sample was partitioned into four solutions (pH 4.0 acetate buffer, pH 4.5 acetate buffer, pH 4.0 PBS buffer, pH 4.5 PBS buffer), with each of the four solutions being subdivided into 16 equal portions. For each set of 16 portions, half were used with pepsin treatment and the other half were used as controls with only buffer added. The added pepsin solution was made from the same buffer and pH combination as the solution that it was being added into. Pepsin was used in a 1:40 pepsin:antibody mass ratio in order to minimize potential damage of the antibody binding site by pepsin. The solutions with added pepsin were incubated at 37 °C for the different time lengths stated earlier. The reactions were stopped with the addition of 2 M Trizma base solution to yield a solution with pH=8.0. Aliquots from each reaction were then prepared in the SDS-PAGE loading dyes and analyzed using SDS-PAGE.
2.4. Kinetic analyses of F(ab)2 reduction
Anti-goat IgG (1 mg/mL) or anti-human IgG1 (4 mg/mL) was dialyzed overnight in Slide-A-Lyzer dialysis cassettes against pH 4.0 acetate buffer (deionized water, 100 mM sodium acetate). The concentration of the antibody solution was determined using UV–vis analysis at 280 nm following dialysis. To generate F(ab)2 fragments, pepsin solution (0.1 mg/mL pepsin in 100 mM sodium acetate buffer, pH 4.0) was added in a mass ratio of 1:40 enzyme:antibody. The resulting solution was incubated at 37 °C for 24 h. The reaction was quenched with 2 M Trizma base solution to yield a solution with pH=8.0. The treated digest was dialyzed overnight against a preparative pH 5.5 reduction buffer (deionized water, 3 mM EDTA, 100 mM sodium chloride, 50 mM sodium citrate tribasic dehydrate, 100 mM sodium tetratborate decahydrate). The concentration of the antibody solution was determined using UV–vis following dialysis.
Antibodies were cleaved in four different experiments, with each experiment performed in triplicate. The first experiment compared the three reducing agents (DTT, MEA, DTBA) at room temperature (22 °C) against polyclonal anti-goat IgG antibodies. The reducing agents were added to the F(ab)2 solutions at a ratio of 2 mM reducing agent to 1 mg/mL F(ab)2. Aliquots of 100 µL and 2 µL were retrieved for future HPLC and SDS-PAGE analyses at the following time points: 5, 10, 20, 40, 90, 180, 300, and 480 min. HPLC aliquots were frozen immediately using liquid nitrogen and stored at -30 °C. SDS-PAGE samples were quenched with a 10-fold excess of iodoacetamide and allowed to react for 30 min before being stored at -30 °C. The second experiment varied three concentrations of reducing agent – 0.5 mM, 1 mM, and 2 mM DTBA to 1 mg/mL of polyclonal anti-goat IgG F(ab)2 – using the same protocol as described above. The third experiment assessed reducing capabilities of DTT, MEA, and DTBA at physiological temperature (37 °C) using the same procedure with the concentrations held at 2 mM reducing agent to 1 mg/mL of polyclonal anti-goat IgG F(ab)2. It should be noted that the experiments at physiological temperature were stopped after the 90 min time point. The final experiment examined the feasibility of cleavage using monoclonal mouse anti-human IgG1 antibodies at room temperature (22 °C). The concentrations of the reducing agents were held at 2 mM reducing agent to 1 mg/mL of monoclonal anti-human IgG1 antibodies.
HPLC analyses were performed shortly after reduction and samples were thawed individually just before each run. Internal standards containing known concentrations of F(ab)2 and Fab׳ fragments were used to allow quantitative analysis of each sample. Samples for SDS-PAGE analyses were thawed, mixed with loading dyes, and ran on 12% SDS-PAGE gels.
2.5. Ellman׳s test for Fab׳ nucleophilic sulfides
Fab׳ fragments generated from the HPLC separation of the samples were reacted with Ellman׳s reagent. Aliquots of 1 mL containing known amounts of Fab fragments were diluted to 3 mL in pH 7.4 phosphate buffer (100 mM sodium phosphate dibasic, 1 mM EDTA). Ellman׳s reagent (10-fold molar excess) was added and the resulting solutions were monitored in a 1-cm UV cell at 25 °C for 15 min. The formation of 2-nitro-5-thio-benzoic acid was quantitatively determined at 412 nm using an extinction coefficient of 14,150 M−1 [27], [31], [32].
3. Results and discussion
3.1. Optimization of the cleavage protocols
Polyclonal rabbit IgG antibodies were chosen for this study as they have been shown to be a good model for the prediction of general antibody behavior, which may be extended to antibodies from other species [8], [33], [34]. However, it was also important to show the varying effects of the reducing agents on antibodies from different species and with differing clonalities. Thus, monoclonal mouse anti-human IgG1 antibodies were also tested against the reducing agents. For both antibodies, the optimization of pepsin cleavage yielded the best results when pepsin was used in pH 4.0 acetate buffer over 24 h. SDS-PAGE analyses indicated that this time, buffer, and pH combination produced no undesirable cleavage byproducts while maximizing the amount of F(ab)2 generated for the polyclonal rabbit anti-goat IgG antibodies. The monoclonal mouse anti-human IgG1 antibodies were more resilient to the pepsin cleavage and very little cleavage occurred before the 24 h mark. After 48 h, there was no apparent F(ab)2 molecules left in solution, indicating undesirable degradation by pepsin. An additional test showing pepsin cleavage at 24, 28, 32, 36, 40, 44, and 48 h also showed that the maximum amount of F(ab)2 was obtained at the 24 h point for the monoclonal anti-human IgG1 antibodies. It should be noted that undesirable byproducts were observed in all of the time points and complete whole antibody cleavage was not achieved in any of the time points for the monoclonal antibodies.
The reduction buffer conditions were chosen based on previous studies by Jiskoot in 1990 and Zheng in 2006 [35], [36]. Antibodies have been found to precipitate at strongly acidic solutions (pH=3.0) and to decompose in alkaline conditions [35]. Thus, it was necessary to chose a reduction buffer that was able to maintain the ionic strength required for proper antibody conformation as well as a pH that the antibodies could tolerate. Deamidation is a common degradation pathway for antibodies and can be minimized using appropriate buffer constituents. Zheng showed more prominent antibody deamidation in both citrate and phosphate buffers at neutral pH. However, at using citrate buffers at pH 5.5, deamidation was minimized [36]. Another common problem that can result in loss of biological activity is protein aggregation, which is influenced by temperature, ionic strength, and time in solution. It has been shown that citrate buffers, particularly those at pH 5.5, exhibit less protein aggregation compared to phosphate buffers [36].
3.2. Kinetic comparisons of DTT, MEA, and DTBA
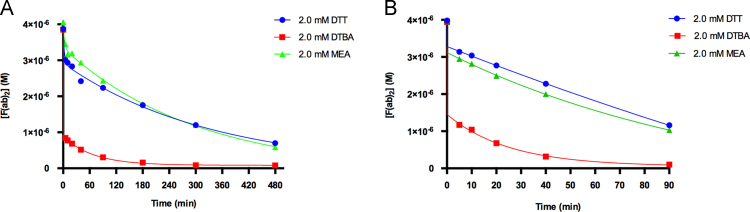
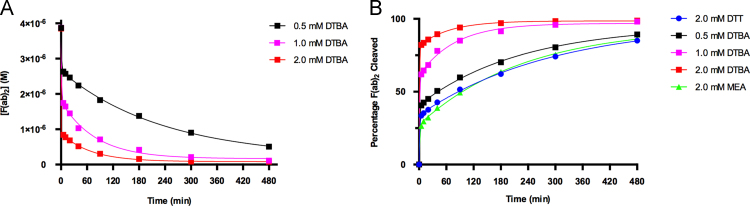
The results of the kinetic analyses following the reduction of F(ab)2 units to Fab׳ fragments are shown in Fig. 3, Fig. 4, Fig. 5. Comparison of the kinetic rate constants of DTT, MEA, and DTBA at room temperature (22 °C) for the polyclonal anti-goat IgG antibodies yields ratios of kDTBA/kDTT≈213, kDTBA/kMEA≈71, and kMEA/kDTT≈3. These ratios indicate kinetic superiority of DTBA in the reduction of F(ab)2 fragments at room temperature. The reduction process was nearly complete in 3 h time using DTBA, whereas both DTT and MEA were far from completion (Fig. 3A). These quantitative results are confirmed using qualitative SDS-PAGE analyses. The SDS-PAGE results (Supporting Information), show a decrease in the intensity of the F(ab)2 band around 110 kDa and an increase in the intensity of the Fab׳ fragment band around 55 kDa. The changes in the two bands are most noticeable in for the 90, 180, and 300 min time points. In these time points, the F(ab)2 band intensity drops off significantly with a sudden increase in Fab׳ band intensity for DTBA reductions. This trend is not observed for the DTT and MEA SDS-PAGE analyses, confirming the kinetic results. There is an increase in the band intensity of the band located near the 26 kDa marker on the SDS-PAGE for the DTT reduction (Fig. S2). This increase in intensity can be attributed to the cleavage of the interchain disulfides between the light and heavy chains of the antibody. This undesirable cleavage is not observed with the DTBA reduction of the polyclonal antibodies, confirmed by both HPLC and SDS-PAGE results.
Fig. 3.
Reduction of polyclonal anti-goat IgG F(ab)2 at (A) room temperature (22 °C) and (B) physiological temperature (37 °C).
Fig. 4.
(A) Reduction of polyclonal anti-goat IgG F(ab)2 at room temperature (22 °C) with varying concentrations of DTBA and (B) formation of polyclonal anti-goat IgG Fab׳ fragments at room temperature with the three reducing agents and the three varying concentrations of DTBA.
Fig. 5.
(A) Reduction of monoclonal anti-human IgG1 F(ab)2 at room temperature (22 °C) and (B) formation of monoclonal anti-human IgG1 Fab׳ fragments.
The relatively small difference in the rate constants between DTT and MEA agree with the observation that both reducing agents are commonly used for the cleavage of F(ab)2 fragments [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. It should be noted that due to the extremely fast reducing capability of DTBA in the first 5 min of cleavage, the rate constant cannot be reliably quantified with the method used in this study and should be interpreted qualitatively with respects to the rate constants of DTT and MEA. These observations should be kept in mind for all three experiments involving DTBA reduction of polyclonal anti-goat IgG antibodies.
The results from the cleavage of F(ab)2 fragments at physiological temperature (Fig. 3B) show an interesting change in the relative rates between DTT, MEA, and DTBA. Rate constant analyses (Supporting Information) yield ratios of kDTBA/kDTT≈15, kDTBA/kMEA≈9, and kMEA/kDTT≈2. The difference between rate constants for DTBA at room temperature and physiological temperature did not significantly change, whereas the differences were increased for DTT and MEA. DTBA is still the more efficient reducing agent at physiological temperature, however, not by as large of a difference as is observed at room temperature.
Kinetic rate constant comparisons for three varying concentrations of DTBA at room temperature (Fig. 4A) produce rate constant ratios of k2.0 mM DTBA/k1.0 mM DTBA≈3, k2.0 mM DTBA/k0.5 mM DTBA≈40, k1.0 mM DTBA/k0.5 mM DTBA≈13. As is expected with a pseudo-first order reaction, an increase in the concentration of DTBA results in an increase in the rate of the reaction. Comparisons of the rate constants for the varying concentrations of DTBA with the rate constants determined for DTT and MEA at room temperature (Fig. 3A) give ratios of k1.0 mM DTBA/kDTT≈67, k0.5 mM DTBA/kDTT≈5, k1.0 mM DTBA/kMEA≈22, and k0.5 mM DTBA/kMEA≈2. All of the tested concentrations of DTBA proved to cleave F(ab)2 fragments faster than the 2.0 mM DTT or 2.0 mM MEA concentrations indicating the kinetic superiority of DTBA with polyclonal anti-goat IgG antibodies.
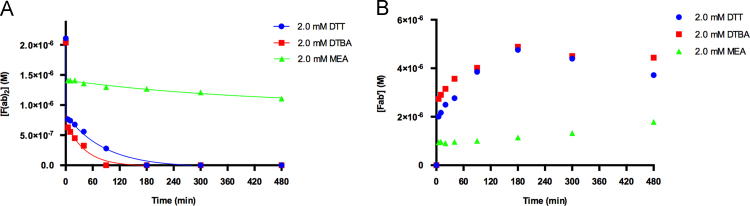
The reduction kinetics for the monoclonal anti-human IgG1 antibodies (Fig. 5A) shows all of the reducing agents to work extremely quickly in the first 5 min of the experiment. Due to this, the calculated pseudo-first order rate constants should be interpreted qualitatively through relative comparison. Comparing the ratios of rate constants shows that kDTBA/kDTT≈2, kDTBA/kMEA≈10, and kDTT/kMEA≈5. Reduction of the F(ab)2 with DTBA and DTT is complete at the 90 and 180 min time points, as confirmed with the SDS-PAGE results. Cleavage of the F(ab)2 with MEA was significantly slower than with the other reducing agents and appeared to have slowed down drastically after the first five minutes. The polyclonal rabbit anti-goat IgG antibodies typically have only one disulfide linkage in their hinge region, whereas it is common to find more than two for monoclonal mouse antibodies [37]. These results show that DTBA and DTT are able to cleave through the tougher hinge region of the mouse antibodies, whereas MEA has a much more difficult time.
It is important to correlate the degradation of the F(ab)2 for both the polyclonal and monoclonal antibodies with the formation of their respective Fab׳ fragments. Cleavage of the polyclonal F(ab)2 with DTT, DTBA, and MEA showed an increase in SDS-PAGE band intensity for the Fab׳ band as the experiment progressed. The increase in Fab׳ fragment concentration was confirmed by HPLC analyses.
This result was similar with the SDS-PAGE band intensities of the monoclonal F(ab)2 reductions with DTT and DTBA. However, very little increase in Fab׳ band intensity was observed for the reduction with MEA. As determined with the HPLC analyses, the concentration of the monoclonal Fab׳ fragments (Fig. 5B) increased significantly during the DTT and DTBA reductions. The HPLC analyses indicated a decrease in Fab׳ fragment concentrations for these two reductions after the 180 min time point. This may have started prior to that time, yet it was not noticed in the HPLC analyses as the rate of Fab׳ fragment formation was greater than the rate of Fab׳ fragment degradation until after the 180 min time point. This result is strengthened by the increase of the 26 kDa band intensity of the SDS-PAGE analyses prior to the 180 min time point. Most likely, DTT and DTBA were participating in interchain disulfide cleavages, resulting in an increase in the concentration of the light chains in solution. These findings show that it is crucial to optimize antibody cleavage in order to maximize on the desired Fab׳ fragments and minimize on undesirable cleavages.
With the production of Fab׳ fragments confirmed, it was necessary to determine whether or not DTBA preserved the nucleophilic C-terminal sulfides on the fragments. Isolated Fab׳ fragments from DTT, MEA, and DTBA cleavages of both polyclonal and monoclonal antibodies were subjected to Ellman׳s reagent [27], [31], [32]. There was no significant difference in the number of nucleophilic sulfides per Fab׳ fragment when the products from DTT, MEA, and DTBA cleavages were compared. The Fab׳ fragments produced by all the reducing agents were found to have between 0.90 and 0.94 nucleophilic sulfides per Fab׳ fragment. This result is similar to that observed with the Fab׳ fragments obtained after the monoclonal antibody reductions. The monoclonal Fab׳ fragments were found to have between 3.66 and 3.82 nucleophilic sulfides per Fab׳ fragment.
In conclusion, the F(ab)2 reduction experiments conducted in this study have shown DTBA to be the more efficient reducing agent compared to DTT and MEA. The high reduction capabilities observed by DTBA in the first 5 min of the experiment do not allow for reliably quantifiable rate constants. Thus, the rate constant analyses should be interpreted qualitatively through the relative comparison of the values. With the cleavage of polyclonal rabbit anti-goat IgG F(ab)2, DTBA was ~213 times faster than DTT and ~71 times faster than MEA. This trend was observed with the cleavage of the monoclonal mouse anti-human IgG1 F(ab)2 as DTBA was ~2 times faster than DTT and ~10 times faster than MEA. Due to the increased number of hinge disulfides in the monoclonal F(ab)2, difficulties with the cleavage were observed with MEA. No problems were detected with the DTT and DTBA reductions. However, DTT and DTBA were both observed to cleave the interchain disulfide bonds of the antibody fragments in the monoclonal antibody reductions. It is very important that antibody cleavage is optimized in order to maximize the yield of the desirable Fab׳ fragments.
Acknowledgments
The authors are grateful to the Natural Sciences and Engineering Research Council of Canada for support of this work.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2015.04.004.
Appendix A. Supplementary materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
References
- 1.Guzman N.A. Improved solid-phase microextraction device for use in on-line immmunoaffinity capillary electrophoresis. Electrophoresis. 2003;24:3718–3727. doi: 10.1002/elps.200305647. [DOI] [PubMed] [Google Scholar]
- 2.Medina-Casanellas S., Benavente F., Barbosa J. Preparation and evaluation of an immunoaffinity sorbent with Fab׳ antibody fragments for the analysis of opioid peptides by on-line immunoaffinity solid-phase extraction capillary electrophoresis-mass spectrometry. Anal. Chim. Acta. 2013;789:91–99. doi: 10.1016/j.aca.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Guzman N.A. Determination of immunoreactive gonadotropin-releasing hormone in serum and urine by on-line immunoaffinity capillary electrophoresis coupled to mass spectrometry. J. Chromatogr. B. 2000;749:197–213. doi: 10.1016/s0378-4347(00)00410-2. [DOI] [PubMed] [Google Scholar]
- 4.Peluso P., Wilson D.S., Do D. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 5.Lu B., Smyth M.R., O׳Kennedy R. Oriented Immobilization of Antibodies and Its Applications in Immunoassays and Immunosensors. Analyst. 1996;121:29R–32R. doi: 10.1039/an996210029r. [DOI] [PubMed] [Google Scholar]
- 6.Vikholm-Lundin I., Auer S., Hellgren A.-C. Detection of 3,4-methylenedioxymethamphetamine (MDMA, ecstasy) by displacement of antibodies. Sens. Actuator B–Chem. 2011;156:28–34. [Google Scholar]
- 7.Neves-Peterson M.T., Snabe T., Klitgaard S. Photonic activation of disulfide bridges achieves oriented protein immobilization on biosensor surfaces. Protein Sci. 2006;15:343–351. doi: 10.1110/ps.051885306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke W., Beckwith J.D., Jackson A. Antibody immobilization to high-performance liquid chromatography supports: characterization of maximum loading capacity for intact immunoglobulin G and Fab fragments. J. Chromatogr. A. 2000;888:13–22. doi: 10.1016/s0021-9673(00)00548-3. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y., Gutshall L., Jiang H. Two routes for production and purification of Fab fragments in biopharmaceutical discovery research: papain digestion of mAb and transient expression in mammalian cells. Protein Exp. Purif. 2009;67:182–189. doi: 10.1016/j.pep.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Roque A.C.A., Taipa M.A., Lowe C.R. Synthesis and screening of a rationally designed combinatorial library of affinity ligands mimicking protein L from Peptostreptococcus magnus. J. Mol. Recognit. 2005;18:213–224. doi: 10.1002/jmr.733. [DOI] [PubMed] [Google Scholar]
- 11.Weber V., Linsberger I., Ettenauer M. Development of specific adsorbents for human tumor necrosis factor-α: influence of antibody immobilization on performance and biocompatibility. Biomacromolecules. 2005;6:1864–1870. doi: 10.1021/bm040074t. [DOI] [PubMed] [Google Scholar]
- 12.Catimel B., Nerrie M., Lee F.T. Kinetic analysis of the interaction between the monoclonal antibody A33 and its colonic epithelial antigen by the use of an optical biosensor: a comparison of immobilization strategies. J. Chromatogr. A. 1997;776:15–30. doi: 10.1016/s0021-9673(97)00087-3. [DOI] [PubMed] [Google Scholar]
- 13.Kumada Y., Hamasaki K., Nakagawa A. Immobilization and functional reconstitution of antibody Fab fragment by solid-phase refolding. J. Immunol. Methods. 2013;400–401:70–77. doi: 10.1016/j.jim.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Le B., Shinkai M., Kitade T. Preparation of tumor-specific magnetoliposomes and their application for hyperthermia. J. Chem. Eng. Jpn. 2001;34:66–72. [Google Scholar]
- 15.Bowles M., Johnston S.C., Schoof D.D. Large scale production and purification of paraquat and desipramine monoclonal antibodies and their Fab fragments, Int. J. Immunopharmac. 1988;10:537–545. doi: 10.1016/0192-0561(88)90071-9. [DOI] [PubMed] [Google Scholar]
- 16.Helali S., Abdelghani A., Hafaiedh I. Functionalization of niobium electrodes for the construction of impedimetric biosensors. Mater. Sci. Eng. C. 2008;28:826–830. [Google Scholar]
- 17.Adlersberg J.B. The immunoglobulin hinge (Interdomain) region. Ric Clin. Lab. 1976;6:191–205. [PubMed] [Google Scholar]
- 18.Edmundson A.B., Guddat L.W., Andersen K.N. Crystal Structures of Intact IgG Antibodies. Immuno Methods. 1993;3:197–210. [Google Scholar]
- 19.Benny A.G., Lint R., Henry S.M. A simple standardized method for the preparation of pure fab fragments of rabbit immunoglobulin G. N. Z. J. Med. Lab. Technol. 1987;41:38–39. [Google Scholar]
- 20.Wilson D.S., Wu J., Peluso P. Improved method for pepsinolysis of mouse IgG1 molecules to F(ab׳)2 fragments. J. Immunol. Methods. 2002;260:29–36. doi: 10.1016/s0022-1759(01)00514-2. [DOI] [PubMed] [Google Scholar]
- 21.Mariani M., Camagna M., Tarditi L. A new enzymatic method to obtain high-yield F(ab)2 suitable for clinical use from mouse IgG1. Mol. Immunol. 1991;28:69–77. doi: 10.1016/0161-5890(91)90088-2. [DOI] [PubMed] [Google Scholar]
- 22.Akita E.M., Nakai S. Production and purification of Fab׳ fragbments from chicken egg yolk immunoglobulin Y (IgY) J. Immunol. Methods. 1993;162:155–164. doi: 10.1016/0022-1759(93)90380-p. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi Y., Kim H., Kato K. Proteolytic fragmentation with high specificity of mouse immunoglobulin G. J. Immunol. Methods. 1995;181:259–267. doi: 10.1016/0022-1759(95)00010-8. [DOI] [PubMed] [Google Scholar]
- 24.Lukesh J.C., Palte M.J., Raines R.T. A potent, versatile disulfide-reducing agent from aspartic acid. J. Am. Chem. Soc. 2012;134:4057–4059. doi: 10.1021/ja211931f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleland W.W. Dithiothreitol, a new protective reagent for SH groups. Biochemistry. 1963;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 26.Burns J.A., Butler J.C., Moran J. Selective Reduction of Disulfides by Tris(2-carboxyethyl)phosphine. J. Org. Chem. 1991;56:2648–2650. [Google Scholar]
- 27.Whitesides G.M., Lilburn J.E., Szajewski R.P. Rates of thiol-disulfide interchange reactions between mono and dithiols and Ellman׳s reagent. J. Org. Chem. 1977;42:332–338. [Google Scholar]
- 28.Gray W.R. Disulfide structures of highly bridged peptides: a new strategy for analysis. Protein Sci. 1993;2:1732–1748. doi: 10.1002/pro.5560021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cline D.J., Redding S.E., Brohawn S.G. New water-soluble phosphines as reductants of peptide and protein disulfide bonds: reactivity and membrane permeability. Biochemistry. 2004;43:15195–15203. doi: 10.1021/bi048329a. [DOI] [PubMed] [Google Scholar]
- 30.Lukesh J.C., VanVeller B., Raines R.T. Thiols and selenols as electron-relay catalysts for disulfide-bond reduction. Angew. Chem. Int. Ed. 2013;52:12901–12904. doi: 10.1002/anie.201307481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaked Z., Szajewski R.P., Whitesides G.M. Rates of thiol-disulfide interchange reactions involving proteins and kinetic measurements of Thiol pKa values. Biochemistry. 1980;19:4156–4166. doi: 10.1021/bi00559a004. [DOI] [PubMed] [Google Scholar]
- 32.Eyer P., Worek F., Kiderlen D. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal. Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe C.A.C., Hage D.S. Studies on the rate and control of antibody oxidation by periodate. Anal. Biochem. 1995;231:123–130. doi: 10.1006/abio.1995.1511. [DOI] [PubMed] [Google Scholar]
- 34.Oates M.R., Clarke W., Marsh E.M. Kinetic Studies on the Immobilization of antibodies to high-performance liquid chromatographic supports. Bioconjug. Chem. 1998;9:459–465. doi: 10.1021/bc970177r. [DOI] [PubMed] [Google Scholar]
- 35.Jiskoot W., Beuvery E.C., de Koning A.A.M. Analytical approaches to the study of monoclonal antibody stability. Pharm. Res. 1990;7:1234–1241. doi: 10.1023/a:1015925519154. [DOI] [PubMed] [Google Scholar]
- 36.Zheng J.Y., Janis L.J. Influence of pH, buffer species, and storage temperature on physicochemical stability of a humanized monoclonal antibody LA298. Int. J. Pharm. 2006;308:46–51. doi: 10.1016/j.ijpharm.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 37.Liu H., May K. Disulfide bond structures of IgG molecules. mAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data