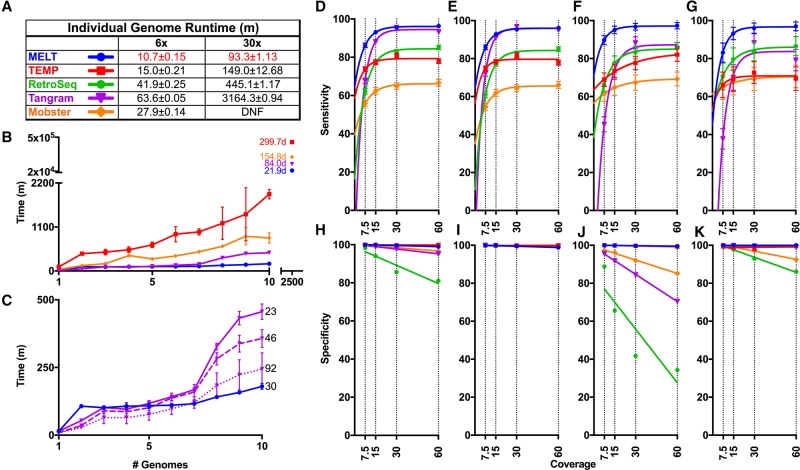
Figure 1.
Comparisons of MEI discovery algorithms. (A–C) Runtime comparisons between MELT and four other MEI discovery algorithms: RetroSeq Mobster, Tangram, and TEMP. (A) Runtime in minutes on either a 6× or 30× coverage genome using a single processor (numbers are mean ± SD), with the best time for each coverage indicated in red. (B) Time required for each algorithm to analyze between one and 10 genomes using a distributed computing cluster. Shown to the right of experimental data are extrapolated estimates of the total runtimes for 2504 genomes with each algorithm. (C) Identical to B but depicting the median runtime for only MELT and Tangram. Tangram was run with 23, 46, or 92 threads (numbers to the right of lines). (D–G) Comparison of sensitivities for MELT and the MEI detection algorithms outlined above. False negative rates (FNRs) are plotted for (D) Aggregate, (E) Alu, (F) L1, and (G) SVA. (H–K) Comparison of specificities for MELT and the MEI detection algorithms outlined above. False discovery rates (FDRs) are plotted for (H) Aggregate, (I) Alu, (J) L1, and (K) SVA (Supplemental Table S5).