Abstract
Large-scale efforts like the ENCODE Project have made tremendous progress in cataloging the genomic binding patterns of DNA-associated proteins (DAPs), such as transcription factors (TFs). However, most chromatin immunoprecipitation-sequencing (ChIP-seq) analyses have focused on a few immortalized cell lines whose activities and physiology differ in important ways from endogenous cells and tissues. Consequently, binding data from primary human tissue are essential to improving our understanding of in vivo gene regulation. Here, we identify and analyze more than 440,000 binding sites using ChIP-seq data for 20 DAPs in two human liver tissue samples. We integrated binding data with transcriptome and phased WGS data to investigate allelic DAP interactions and the impact of heterozygous sequence variation on the expression of neighboring genes. Our tissue-based data set exhibits binding patterns more consistent with liver biology than cell lines, and we describe uses of these data to better prioritize impactful noncoding variation. Collectively, our rich data set offers novel insights into genome function in human liver tissue and provides a valuable resource for assessing disease-related disruptions.
Complex gene regulatory networks underlie key aspects of human development, tissue physiology, and cell fate determination (Karlebach and Shamir 2008; Spitz and Furlong 2012). These gene expression programs are coordinated by DNA-associated proteins (DAPs), especially sequence-specific transcription factors (TFs), which bind to promoters, enhancers, silencers, insulators and other cis-regulatory elements (Spitz and Furlong 2012). Owing to their fundamental biological importance, disease can result from disruption or alteration of trans-acting DAPs or the cis-regulatory elements to which they bind (Khurana et al. 2016; Sur and Taipale 2016). Accordingly, the interactions of DAPs and cis-regulatory sequences have been investigated extensively (The ENCODE Project Consortium 2007, 2012; Gerstein et al. 2012; Andersson et al. 2014). These studies have been greatly aided by high-throughput sequencing technologies that map genome-wide binding patterns of DAPs, in particular via chromatin immunoprecipitation sequencing (ChIP-seq) (Johnson et al. 2007; Robertson et al. 2007).
The vast majority of genome-wide DAP binding maps, including many generated by the ENCODE Project (https://www.encodeproject.org), are based on a small number of mostly tumor-derived cell lines. These studies revealed strong correlations between open chromatin, transcription factor binding, DNA methylation levels, and transcription of nearby genes (Xie et al. 2013). Studies of DNA binding proteins reveal regulatory networks associated with a variety of experimental perturbations and some cell-type specificity (Gerstein et al. 2012; Gertz et al. 2012a; Reddy et al. 2012a; Savic et al. 2015a, 2016). Additional research further explored the molecular determinants that contribute to cell-type specificity (Gertz et al. 2013; Mortazavi et al. 2013), including the identification of sequence variants that drive changes to the epigenome (McVicker et al. 2013) and genomic hallmarks that predict active TF binding sites (Savic et al. 2015b). However, these in vitro systems are likely to be limited in the extent to which they recapitulate in vivo tissue environments, especially for noncancerous tissues (Sandberg and Ernberg 2005; Ertel et al. 2006).
The generation of genome-wide, DAP binding patterns in healthy human tissue is essential to improving our understanding of transcriptional control within a physiological context. Recently, reference epigenomes, consisting of histone modification, DNase hypersensitivity, and DNA methylation measurements, have been compiled for more than 100 primary tissues by the Roadmap Epigenomics Consortium et al. (2015). Their integrative analysis found that regulatory marks, particularly enhancer-associated H3K4me1 peaks, exhibited tissue-specific enrichment for relevant complex trait–associated variants. Further, epigenomic features were found to be highly predictive of cancer somatic mutation burdens, and the chromatin landscape of the appropriate primary tissue greatly outperformed that of matched cancer cell lines (Polak et al. 2015). These findings demonstrate the promise of mapping the regulatory landscape of primary tissues and highlight a need for identifying the trans-acting factors bound to likely regulatory regions and the sequence variation that may affect their activity.
Results
DAPs display extensive colocalization
We performed pairs of replicate ChIP-seq assays for each of the 20 DAPs (Supplemental Table S1) in two primary liver tissue samples (Supplemental Table S2), for a total of 80 ChIP-seq experiments. We selected DAPs based on the availability of suitable ChIP-seq-grade antibodies and their expression levels in the liver, ultimately assaying 17 sequence-specific TFs, two DAPs involved in maintaining chromatin structure, CTCF and RAD21, and RNA polymerase II (POLR2A), which is directly involved in transcription. ChIP-seq experiments were conducted in accordance with ENCODE guidelines (Landt and Marinov 2012). All replicate pairs were strongly correlated, and canonical motif enrichment was detected for all sequence-specific TFs (Supplemental Table S3; Supplemental Material). We identified between 909 and 60,597 binding events for each DAP and tissue sample (Supplemental Table S1) and identified more than 440,000 binding sites, spread over 150,000 unique genomic locations, across all DAPs in both livers.
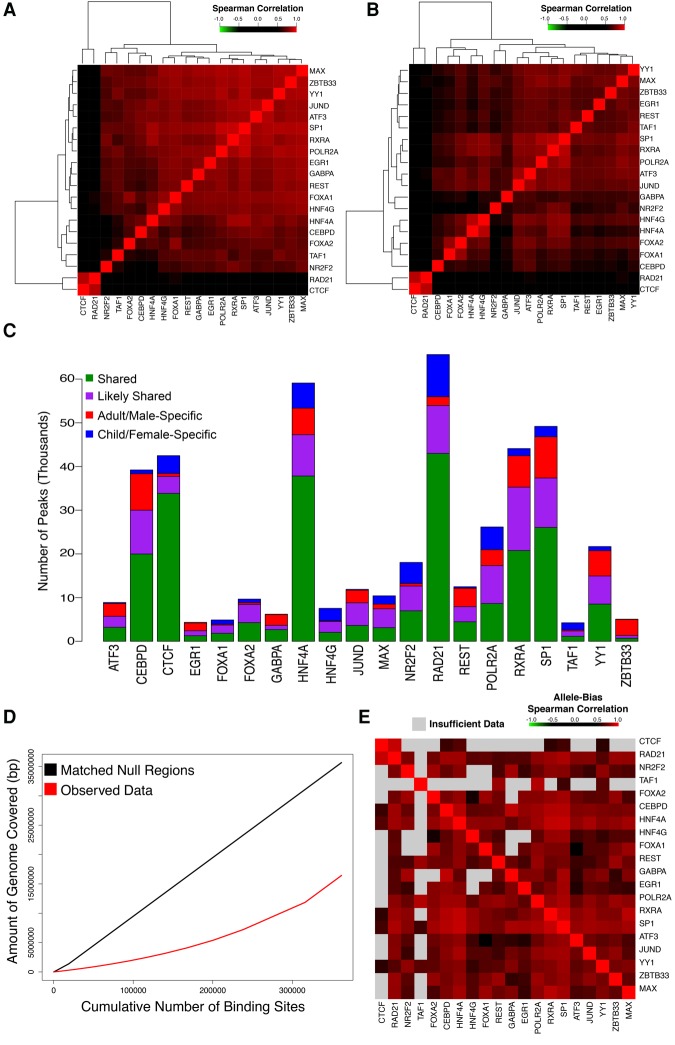
As others have previously observed in cell lines (Xie et al. 2013), we found a high degree of colocalization among our assayed factors. Hierarchical clustering of normalized ChIP-seq read count correlations revealed strong correlations between many pairs of DAPs in both tissues (median rho of 0.718 and 0.642, and maximum rho of 0.889 and 0.863, in the adult/male and child/female livers, respectively), with no factors negatively correlated (Fig. 1A,B). We further found that, despite age, sex, and other life history differences, ∼75% of all DAP binding sites were shared between the two donors in at least one replicate (53% are common among all four data sets) (Fig. 1C). We observed stronger binding similarity between maps of a given DAP from the two samples than between two different DAPs in the same sample (Wilcoxon test, P < 0.0001) (Supplemental Figs. S1, S2). Consistent with their roles in maintaining genome insulation and three-dimensional genome structure (Merkenschlager and Nora 2016), RAD21 and CTCF displayed the most distinctive binding patterns and clustered separately from all other DAPs in both tissues. To determine whether the degree of interaction we observed between factors exceeds random expectation, we randomly sampled genomic regions matched for length, GC content, and repetitive sequence content to the observed binding sites (Fletez-Brant et al. 2013). Compared to randomly sampled regions, observed DAP binding sites covered ∼50% fewer bases (Fig. 1D), indicating that observed overlap rates are far above random expectation (P < 0.0001). Binding sites of FOXA1, a pioneer factor (Zaret and Carroll 2011), had the greatest mean number of overlapping sites, and the degree of colocalization at FOXA1 binding sites differed dramatically from non-FOXA1 bound sites (Supplemental Fig. S3).
Figure 1.
DAPs exhibit extensive binding colocalization. (A,B) Heatmaps of Spearman correlation matrix of normalized DAP binding intensities at all observed binding sites in the adult/male (A) and child/female (B) liver. (C) Stacked bar plot displaying the number of peaks for each TF. Bars are divided into those that are shared between both replicates of both donors (green), shared between a donor and one replicate of the other donor (purple), or specific to the adult/male (red) or child/female donor (blue). (D) Cumulative number of base pairs covered per binding site included in adult/male liver observed data (red) and null regions (black) matched for length, GC content, and repeat content. (E) Heatmap of a pairwise Spearman correlation matrix, ordered identically to A, indicating the correlation of allele bias in DAPs from the adult donor that overlap a heterozygous SNP for each pair of factors. The color of each panel indicates the strength of the correlation, with gray indicating that less than 25 peaks met inclusion criteria for allele bias analysis for a given pair.
We used phased WGS data to examine the allele specificity of DAP colocalization. We assessed the degree of allele bias, measured as the fraction of ChIP-seq reads containing the reference sequence (hg19), for each DAP at all heterozygous single-nucleotide variants (SNVs) that overlapped with an adult DAP binding site (DeSantiago et al. 2017). Correlation analyses of the degree of allele bias at each SNV between all possible pairs of DAPs revealed that factors possessed highly correlated allele biases, indicating that groups of DAPs bound near one another show preference for the same allele (Fig. 1E; Supplemental Table S4). Our binding sites are derived from bulk tissue; thus, DAP binding hubs may arise from direct DAP–DAP interactions or reflect open chromatin where multiple factors are capable of binding.
There is significant interest in using chromatin modifications and DAP binding to identify phenotypically relevant regulatory sites, and previous work demonstrated that clusters of DAP binding sites can be useful predictors of enhancer activity (Dogan et al. 2015). We found that sites bound by more DAPs were more strongly conserved across the mammalian phylogeny (rho = 0.960, P = 7.08 × 10−6) (Supplemental Fig. S4A). These sites were also enriched for the activating histone 3 acetylation marks on lysine 9 (H3K9ac) and lysine 27 (H3K27ac), and they were relatively depleted for the repressive histone 3 methylation marks on lysine 9 (H3K9me3) and lysine 27 (H3K27me3) (Supplemental Fig. S4B–E). Together, these data show that DAPs colocalize extensively, often at the same allele, and sites of increased DAP interaction occur in evolutionarily conserved regions of open chromatin.
DAP binding recapitulates known liver expression programs
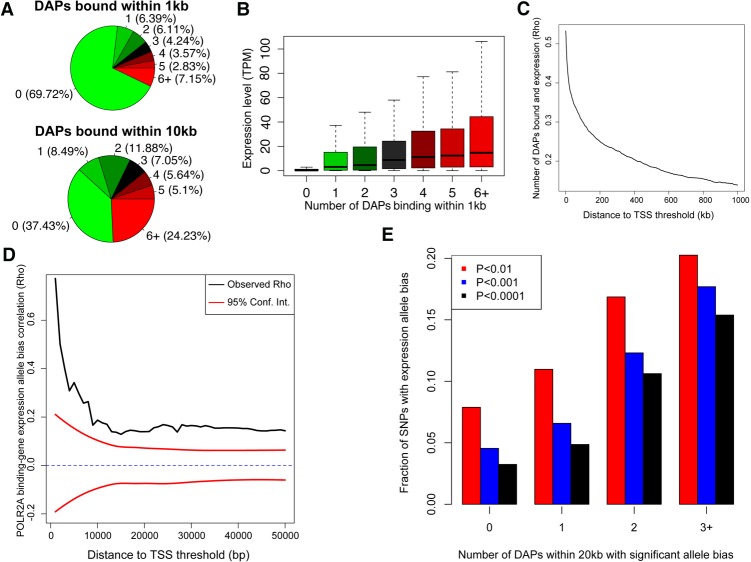
A catalog of DAP binding in liver tissue allows for consideration of the functional consequences of binding on gene expression. We performed quadruplicate RNA-seq experiments on each donor tissue. In general, gene expression was correlated between livers, but genes with an expression change of fourfold or greater overlapped significantly with genes associated with age and gender in an independent liver tissue data set (Fisher's exact test, P = 3.3 × 10−125 and 0.031) (Supplemental Fig. S5; The GTEx Consortium 2015). Integrating expression data with DAP binding, we found that 30% of all Ensembl (GRCh37_E75) annotated genes harbored at least one binding site within 1 kb of their transcriptional start sites (TSSs), and >7% of genes harbored binding events for six or more different DAPs in both adult and child (Fig. 2A; Supplemental Fig. S6A). POLR2A promoter binding within 1 kb of a gene's TSS was strongly associated with expression in both livers (Wilcoxon test, P < 0.0001, mean TPM without POLR2A = 22.3, and mean TPM with POLR2A = 121.8). Gene expression level was also strongly correlated with the number of factors bound within 1 kb of their TSS (rho = 0.533 for adult and rho = 0.526 for child) (Fig. 2B; Supplemental Fig. S6B), however, this effect diminished rapidly as the distance to TSS expanded beyond 1 kb (Fig. 2C).
Figure 2.
DAP occupancy correlates with gene expression. (A) Pie chart representing the percentage of genes containing a specified number of bound DAPs within 1 kb of their TSS. (B) Expression level of genes binned by the number of DAPs bound within 1 kb of their TSS in the adult/male donor. (C) Correlation between expression level of genes and the number of factors bound (as described in B) for a range of distance to TSS thresholds. Correlations are aggregated cumulatively. (D) Correlation between POLR2A binding and neighboring gene expression allele bias over a range of distance thresholds. Ninety-five percent confidence intervals (red) were calculated by randomly shuffling all SNP pairs that met a distance threshold 100 times. (E) Bar plot showing the fraction of expressed SNPs with significant allele bias with varying numbers of neighboring DAPs that also exhibited allele bias within 20 kb.
Based on the allele bias in DAP binding, we expected a similar bias in gene expression with preference to bound alleles. Using phased WGS data with RNA-seq, we found bias in POLR2A occupancy within 1 kb of a gene's TSS was strongly correlated (rho = 0.750) with expression of the same allele. The strength of this correlation dropped rapidly as the distance between binding site and TSS was expanded (Fig. 2D). A similar pattern was observed with several other DAPs (e.g., CEBP, MAX, RXRA and SP1), although the strength of these correlations was reduced and many correlations fell within the null expectation at a majority of distance thresholds. Conversely, repressive factors such as REST and NR2F2 exhibited negative correlations (Supplemental Figs. S7, S8; Supplemental Table S5). EGR1 also displayed a negative correlation with allele expression. EGR1 is not generally considered to be a repressive factor, although a few studies demonstrated potent EGR1 repressive activity (Tan et al. 2003; Feng et al. 2015). The number of neighboring DAPs with significant allele bias was also associated with the likelihood of observing significant allele bias in gene expression (Fig. 2E).
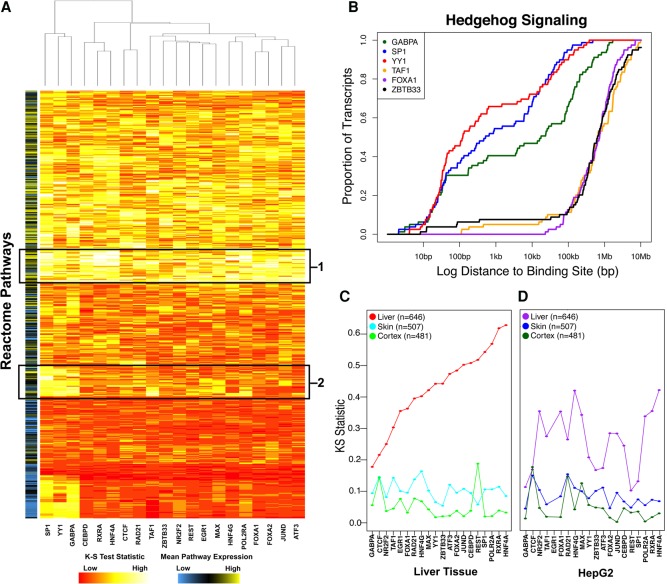
We also explored the consequences of DAP binding by identifying the pathways that might be regulated by these factors. For each DAP, we calculated the distance between the TSSs of genes in every Reactome (http://www.reactome.org) pathway and the nearest binding site for that DAP. We compared the distribution of those distances to that of the background transcriptome using a Kolmogorov–Smirnov (KS) test (Fig. 3A; Supplemental Fig. S9; Supplemental Table S6). For nearly all DAPs (median KS test P < 0.05), we observed strong enrichments for pathways highly active in liver tissue such as lipid and carbohydrate metabolism, drug metabolism, and complement activation (indicated by “1” in Fig. 3A). We identified other pathways specific to subsets of DAPs. Pathways regulating stem-cell state and cell division (indicated by “2” in Fig. 3A) were largely restricted to SP1, YY1, and GABPA binding events. For example, the Hedgehog “on” state (REACT_268718) pathway acts as a key regulator of animal development and differentiation (Ingham et al. 2011). SP1, YY1, and GABPA were all bound within 1 kb of the TSSs of nearly 50% of the 60 genes within this pathway. In comparison, a distance threshold of 1 Mb is required to achieve a similar degree of occupancy for any other DAP (Fig. 3B; Supplemental Fig. S10). SP1, YY1, and GABPA have been previously described as interacting partners (Galvagni et al. 2001; Rosmarin et al. 2004), and these pathway enrichments are consistent with previous studies implicating GABPA in controlling stem cell maintenance and differentiation (Yu et al. 2016).
Figure 3.
Primary liver tissue data recapitulate liver expression programs. (A) Heatmap of KS-test statistic indicating the level of enrichment for proximal binding to each Reactome pathway for each DAP. The color bar on the left indicates the mean expression level of genes within a pathway. Boxed regions indicate core liver pathways bound by all DAPs (1) or DAP-specific pathways involved in cell division and differentiation (2). (B) Representative private pathway plot demonstrating enrichment for proximal GABPA (green), SP1 (blue), and YY1 (red) binding compared to TAF1 (orange), FOXA1 (purple), and ZBTB33 (black). (C,D) Dots represent KS-test statistic of enrichment for proximal binding of each factor to liver (red/purple), skin (blue), and cortex (green)-specific genes in adult/male tissue (C) and HepG2 cells (D).
An important metric by which to assess the utility of our data set is its enrichment for liver-specific gene programs relative to cell line–derived DAP occupancy data. We defined liver-specific genes as those with a mean RPKM of at least two across all GTEx liver tissues and at least fivefold higher than the mean RPKM in all other nonliver tissues. HNF4A and RXRA showed the greatest enrichment for binding near the TSSs of liver-specific genes (Fig. 3C; Supplemental Fig. S11), underscoring their importance in regulating liver-specific functions (Martinez-Jimenez et al. 2010; DeLaForest et al. 2011; Li et al. 2015). Conversely, skin and cortex-specific genes exhibited a much lower enrichment for proximal DAP binding compared to liver-specific genes. Notably, a similar analysis of ChIP-seq data generated by our group in HepG2 cells also revealed enrichment for promoter-proximal binding to liver-specific genes (Fig. 3D). However, only 16% of HepG2 peaks are shared with the adult donor; aggregated across all 20 DAPs, proximal binding enrichment was significantly more pronounced in primary liver tissue (paired Wilcoxon test, P = 1.29 × 10−4). Only NR2F2 and HNF4G exhibited even nominally greater proximal binding enrichment in HepG2 cells than in liver tissue (Fig. 3D). These results were robust to different thresholds used to define tissue specificity (Supplemental Fig. S12). Previous analysis of open chromatin across cell lines has been successful in predicting cell-type–specific expression (Natarajan et al. 2012), and these results suggest similar approaches utilizing DAP occupancy could also be fruitful.
DAP binding sites are enriched for expression-QTL SNPs
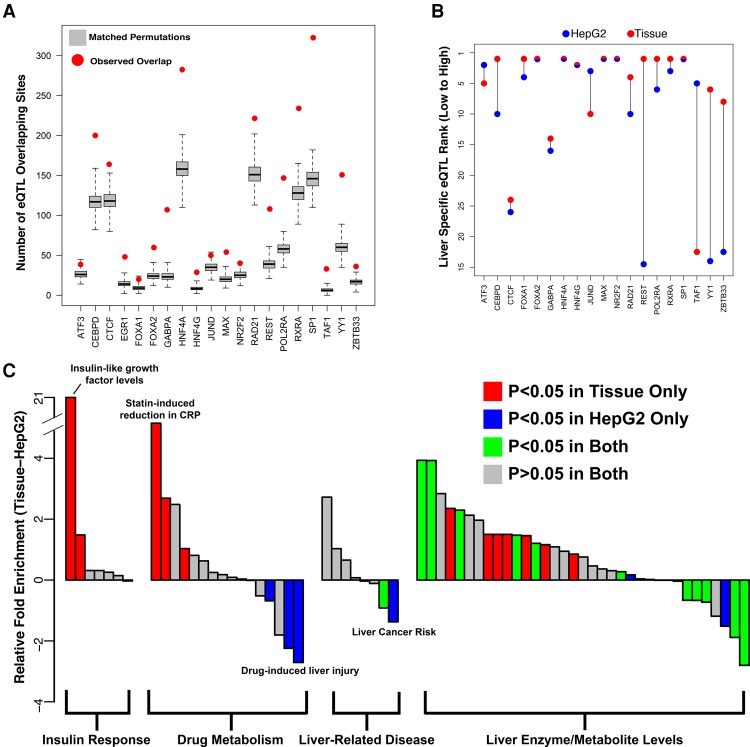
We tested whether DAP binding sites, due to their demonstrated effects on gene expression, were enriched for expression-QTL (eQTL) SNPs in liver tissue cataloged by the GTEx Project. We compared the number of significant eQTL SNPs overlapping binding sites for a given DAP to 1000 randomly sampled sets of SNPs that passed GTEx filtering, controlling for distance to nearest TSS and minor allele frequency. This analysis revealed significant enrichment (Bonferonni-adjusted P < 0.05) for 18 of the 20 assayed DAPs (Fig. 4A; Supplemental Table S7A); the remaining two, JUND and ATF3, trended toward significance (Bonferonni-adjusted P = 0.06 and 0.12). This enrichment was generally specific to liver eQTL SNPs. Repeating the analysis on three tissues (uterus, vagina, anterior cingulate cortex) with <35% eQTL SNPs shared with liver revealed significantly less enrichment (Supplemental Table S7B). To examine tissue specificity more comprehensively, we assessed DAP binding site overlap with tissue-specific eQTLs (FDR < 0.05 in only one tissue). Our DAP binding sites overlapped with liver-specific eQTLs more often than any other tissue for 11 of 20 DAPs. (Supplemental Fig. S13; Supplemental Table S8).
Figure 4.
DAP occupancy is enriched for relevant trait- and expression-associated sequence variation. (A) Red dots indicate the number of eQTLs falling within a DAP binding site relative to the gray box plots, which represent 1000 randomly sampled null SNPs matched for distance to TSS and minor allele frequency. (B) Relative rank of liver-specific eQTL compared to all GTEx tissue specific eQTLs in DAP binding sites assayed in HepG2 cells (blue) and adult liver tissue (red). (C) Difference in enrichment (delta Fisher's exact test odds ratio) for SNPs associated with liver-related phenotypes between binding sites for 19 common DAPs assayed in HepG2 cells and liver tissue. GWAS terms represented by bars are provided in Supplemental Table 9A.
In HepG2 cells, we observed a similarly strong enrichment for liver eQTL SNP overlap for all factors except SP1 (Supplemental Fig. S14A; Supplemental Table S7A). However, this enrichment was much less specific to liver eQTL SNPs, as we observed a stronger enrichment in uterine, vaginal, and anterior cingulate cortex eQTLs (Supplemental Table S7C). Moreover, we observed a reduction in the level of enrichment for liver-specific eQTLs relative to non-liver tissue-specific eQTLs in 10 of 19 DAPs that were assayed in HepG2 cells, suggesting that, at least for some DAPs, tissue ChIP-seq data better identify regions regulating tissue-specific gene expression (Fig. 4B; Supplemental Fig. S14B).
We also identified DAP binding sites that overlap with functional SNPs with liver-related phenotypes by the NHGRI-GRASP genome-wide association study (GWAS) catalog (https://grasp.nhlbi.nih.gov/Overview.aspx). We found greater enrichment (paired Wilcoxon P = 1.8 × 10−3) in liver tissue binding sites for a majority of GWAS terms (45 of 66), including insulin-like growth factor levels and response to statins (Fig. 4C; Supplemental Table S9). HepG2 binding sites did show greater enrichment for a few terms, such as liver cancer risk and HDL cholesterol levels.
DAP binding analyses to prioritize impactful noncoding variation
One of the promises of high-throughput cataloging of DAP binding is better identification of noncoding variation with phenotypically relevant regulatory effects. A challenge associated with using ChIP-seq data for this purpose is that DAP occupancy peaks are often broad, and it is unclear which bases within a ChIP-seq peak significantly affect DAP binding. We therefore assessed the degree of mammalian evolutionary sequence conservation within ChIP-seq peaks using Genomic Evolutionary Rate Profiling (GERP) scores (Cooper et al. 2005). The mean GERP-RS score of each protein's binding sites was significantly greater than the genome-wide average but lower than protein-coding exons (Supplemental Fig. S15). Although these data likely reflect reduced constraint in noncoding sequence relative to coding, ChIP-seq-defined DAP binding sites often do not have the resolution to identify the most critical nucleotides for DAP binding, such as TF motifs, which are often more highly conserved than surrounding sequences (The ENCODE Project Consortium 2012; Reddy et al. 2012b). Recent studies have also indicated that sequence elements critical for determining DAP binding do not necessarily reside solely in the canonical DNA sequence motif (Reddy et al. 2012b; Deplancke et al. 2016; Tehranchi et al. 2016).
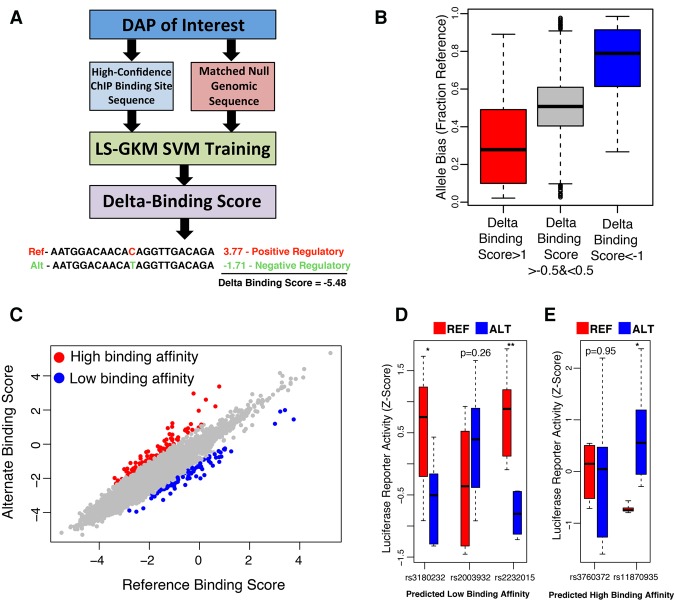
Consequently, to systematically examine evolutionary conservation at base pairs most critical for DAP binding, we applied a previously described method (Lee 2016) to train 10-mer-based support vector machines (SVMs) capable of distinguishing binding sites identified in DAP ChIP-seq experiments from unbound genomic loci matched for GC and repeat content (Fig. 5A). These SVMs were successful in predicting binding for all factors with a mean receiver-operator characteristic area under the curve (ROC-AUC) of 0.928 and precision recall area under the curve (PR-AUC) of 0.702 (Supplemental Fig. S16A,B). A subset of 10-mers, each occurring in a small percentage of total binding sites, were most predictive of DAP binding (Supplemental Fig. S16C). Previous analyses (Xie et al. 2013) identified a relative depletion in DAP motifs at regions of high co-occupancy. We found associations between co-occupancy and SVM classifier scores to be DAP-specific. For CTCF and GABPA, we observed a decrease in our model's confidence in identifying binding at high occupancy sites, but this trend was absent in other DAPs such as HNF4A and RXRA (Supplemental Fig. S17). We computed a “delta” binding score for all possible point mutations within DAP binding sites, defined as the mean decrease in our SVMs’ classifier value for the alternate base relative to the reference sequence. This strategy is similar to a previously developed approach, “deltaSVM,” that focuses on local disruptions of 10-mer feature weights (Lee et al. 2015). Bases with the most negative delta binding score tended to be the most highly conserved for most DAPs (Supplemental Table S10). DAPs with relatively low mean binding site GERP scores, such as GABPA and CTCF, harbored high levels of conservation at their predicted most vulnerable nucleotide positions. We also observed a modest, but significant, correlation between delta binding scores and the observed degree of allele bias (FDR < 0.05) in binding sites for 13 of 20 DAPs (Fig. 5B; Supplemental Fig. S18; Supplemental Table S11), further supporting our confidence in predicting putatively impactful variation at DAP binding sites.
Figure 5.
SNPs capable of disrupting regulatory activity can be identified from liver-derived data. (A) Diagram representing the pipeline for generating SVMs capable of distinguishing DAP binding sites from matched null regions and scoring the predicted impact of all possible mutations on each DAP. (B) Box plots of CTCF binding site overlapping, heterozygous SNPs predicted to be in the top ∼1% for decreasing binding affinity (red), to be in the top ∼1% for increasing binding affinity (blue), and to have no significant impact on binding affinity (gray). The y-axis indicates the fraction of ChIP-seq reads mapping to the reference allele. (C) SVM scores for reference and alternate GTEx liver eQTL SNP alleles for CTCF. Red dots indicate SNPs that hold a positive delta binding score in the top 0.1 percentile. Blue dots indicate SNPs that hold a negative delta-binding score that falls in the bottom 0.1 percentile of all scores. (D,E) Box plots representing the luciferase activity of reference (red) and alternate (blue) sequence in eQTL SNPs predicted to inhibit DAP binding (D) and induce TF binding (E). (*) Two-tailed t-test P < 0.05; (**) P < 0.005.
We ranked all GTEx liver eQTL SNPs using delta binding scores to identify those most likely to alter a DAP binding site (Fig. 5C). Because identifying common sequence variation with functional significance is challenging for eQTL analyses and genome-wide association studies (GWASs) (Edwards et al. 2013), weighting SNPs based on their likelihood to impact DAP binding could be a useful approach for prioritizing follow-up of SNP associations. Of the top 0.1% of DAP-disruptive eQTL SNPs, several were associated with one or more relevant phenotypes described in the NHLBI-GRASP GWAS catalog (Supplemental Tables S12, S13), and they were significantly enriched (Fisher's exact test, P < 0.05) for liver-related GWAS catalog terms compared to all significant liver eQTL SNPs (Supplemental Fig. S19). To validate DAP binding disruptions, we selected five SNPs predicted to either increase or decrease binding affinity and tested them in a luciferase reporter assay in HepG2 cells (Fig. 5D,E; Supplemental Table S14). Predicted SNP effects were confirmed for three of the five SNPs tested (two SNPs with predicted loss of binding and one with greater binding affinity relative to reference sequence), confirming the regulatory impact of these SNPs. Of particular interest is the SNP rs11870935, whose alternate allele is predicted to increase RXRA binding affinity compared to the reference allele. It was associated with cardiovascular disease, LDL cholesterol, and circulating triglyceride levels in a recent GWAS (Teslovich 2010) and was characterized as an intronic/promoter liver eQTL SNP for KPNB1, a gene encoding an importin beta subunit critical for nucleocytoplasmic transport regulating cholesterol biosynthesis and insulin resistance via SREBP and the NF-kB complex, respectively (Nagoshi and Yoneda 2001; Wang et al. 2015).
Disruption of DAP activity in hepatocellular carcinoma
DAP binding is important for the maintenance of tissue identity through regulation of tissue-specific genes. This maintenance is often disrupted during tumorigenesis, facilitating reversion to less differentiated and more proliferative cell states (Sur and Taipale 2016). We integrated our binding data with gene expression data from The Cancer Genome Atlas Project (TCGA; https://cancergenome.nih.gov) to determine the extent to which these factors regulate genes differentially expressed in cancer. We found that DAP binding sites are enriched near genes differentially expressed (FDR < 0.001, DESeq2) (Love et al. 2014) in tumor tissue compared to adjacent normal tissue. In particular, we observed an enrichment for genes down-regulated in tumor tissue (Fig. 6A). HNF4A exhibited the strongest enrichment for binding near genes down-regulated in cancer, in agreement with previous observations implicating it as a tumor suppressor gene (Ning et al. 2010; Bonzo et al. 2012). GABPA and SP1 showed strong enrichment (FDR < 0.05) for binding near genes up-regulated in tumor tissue. This may be due to the importance of GABPA and SP1 proteins for regulating stem cell state and cell division, as described in the Reactome pathway enrichment analysis described above (Fig. 3A).
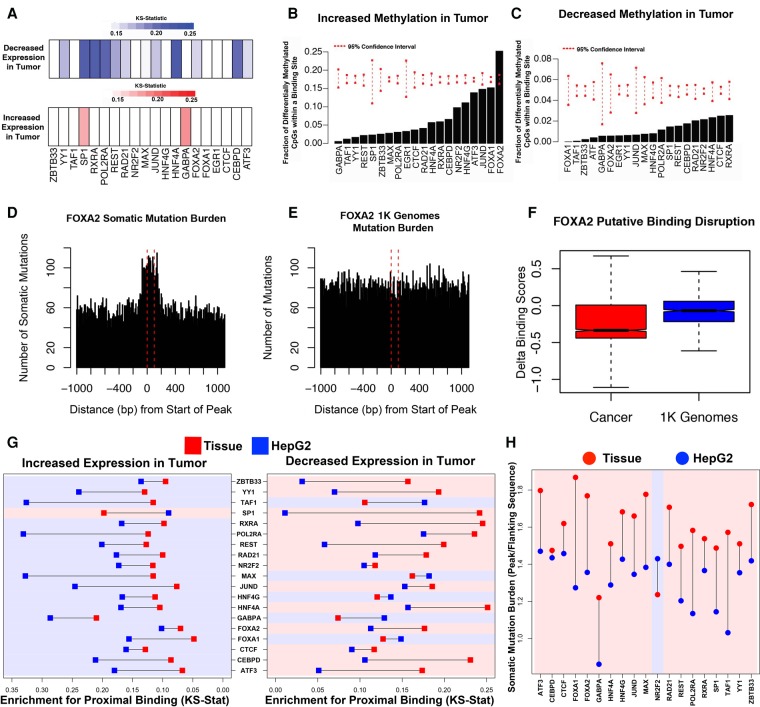
Figure 6.
Primary tissue-derived DAP occupancy complements existing cell line data in characterizing liver cancer–associated genomic changes. (A) Color bars indicating the KS-test statistics for enrichment for binding proximal to the TSS of genes with significantly decreased (blue) or increased (red) expression in tumor tissue compared to adjacent normal tissue for each DAP. (B,C) Percentage of probes with significantly increased or decreased methylation in tumor compared to adjacent normal tissue overlapping a binding site of each DAP. Red dashed lines indicate 95% confidence intervals based on random sampling of an equivalent number of null probes. (D,E) Bars representing the number of somatic mutations (D) or matched 1000 Genome Project (E) mutations observed at contiguous 10-bp bins covering all FOXA2 peaks and flanking 1-kb regions. (F) Delta binding scores of all binding sites overlapping either somatic mutations found in cancer (red) or natural variation measured in 1000 Genomes (blue). (G) KS-test statistics for binding proximal to the TSS of genes with significantly increased or decreased expression in tumor tissue compared to adjacent normal tissue in HepG2 cells (blue) and adult liver tissue (red). (H) Mean somatic mutation burden of DAP binding sites over flanking regions in HepG2 cells (blue) and adult/male tissue (red). Regions are shaded according to whether HepG2- or tissue-derived peaks display greater enrichment.
DNA methylation at DAP binding sites showed a relative depletion in significant differences between tumor and adjacent normal tissue (Fig. 6B,C). However significant increases in methylation levels were more common at binding sites than significant decreases for all DAPs, except GABPA, in agreement with gene expression observations in Figure 6A. FOXA1, which is known to be methylation sensitive (Bartke et al. 2010; Zhu et al. 2016), was the only DAP whose binding site exhibited a significant enrichment for differential methylation. Nearly 25% of FOXA1 binding sites overlapping CpG dinucleotides had significantly increased methylation in tumor compared to adjacent normal tissue (FDR < 0.05).
We also examined somatic variation at all DAP binding sites. Analysis of whole-genome somatic single-nucleotide variation (SNV) data from 258 hepatocellular carcinoma patients obtained from the International Cancer Genome Consortium (ICGC; http://icgc.org) revealed dramatically increased somatic mutation burden in DAP binding sites compared to flanking regions for the majority of tested DAPs (Fig. 6D; Supplemental Fig. S20). This effect was not observed in an equivalent sample of variants from the 1000 Genomes Project matched for reference and alternate base pair composition (1K Genomes; http://www.internationalgenome.org) (The 1000 Genomes Project Consortium 2015) (Fig. 6E). A significant proportion of this increased mutation burden can be explained by mutation rates at cytosine and guanine nucleotides (Supplemental Fig. S21), which are enriched in DAP binding sites (Kaiser et al. 2016). FOXA1 and FOXA2 binding sites demonstrated increased somatic mutation burden after correcting for base-pair composition (P = 0.01 and P = 0.03, respectively), but the robustness of this observation is unclear given that these effects did not survive correction for the scope of hypothesis testing (FDR = 0.285) (Fig. 6D; Supplemental Fig. S20; Supplemental Table S15). Interestingly, mutations in FOXA2 binding sites identified in tumor tissue harbored a more negative delta binding score, on average, compared to binding sites overlapping 1K Genomes SNVs (Wilcoxon P = 6.4 × 10−38) (Fig. 6F).
Our tissue-based DAP binding exhibited increased enrichment for proximal binding near genes down-regulated in liver cancer compared to adjacent normal tissue (paired Wilcoxon P = 4.6 × 10−3), and conversely HepG2-derived DAP binding sites showed greater enrichment for proximal binding near up-regulated genes (paired Wilcoxon P = 2.1 × 10−4) (Fig. 6G). Moreover, HepG2-derived binding sites exhibited a lower somatic mutation burden relative to flanking regions for all factors except for NR2F2 (paired Wilcoxon P = 3.8 × 10−5) (Fig. 6H). More than twice as many NR2F2 peaks were identified in HepG2 relative to our adult male donor, and NR2F2 has an exceptional number of interacting partners that control activity depending on cellular context (Litchfield and Klinge 2012). Therefore, it is perhaps not surprising to see NR2F2 as an outlier in this context. Overall, these data demonstrate that DAP binding sites harbor an increased number of somatic mutations compared to flanking regions, and although a majority of this trend can be attributed to nucleotide composition-related tumor mutational processes, disruption of DAP binding sites may be an important mechanism for altering normal liver gene expression programs. Furthermore, these results suggest noncancerous tissue-based ChIP-seq assays may provide insight to regulatory regions disrupted in cancer that are not accessible from tumor-derived cell lines.
Discussion
We provide a comprehensive evaluation of DAP binding by generating 80 independent ChIP-seq data sets from liver tissues from two individuals. Some of our observations mirrored what has been seen in cell culture–based assays (Yan et al. 2013). For example, we observed a high degree of co-occupancy from our DAPs, including >7% of genes with more than six DAPs bound (Figs. 1C, 2A). DAP co-occupancy showed strong allele bias and correlated with conservation, marks of open chromatin, and neighboring gene expression (Fig. 2B; Supplemental Fig. S4). It remains unclear whether these “hubs” of DAP activity are mediated through direct DAP–DAP interactions or simply facilitated by an open chromatin state established by pioneer factors. Previous investigations (Xie et al. 2013) of genomic regions enriched for hundreds of DAP binding sites found a relative depletion of DAP motifs, implicating nonspecific chromatin accessibility as the driver of promiscuous binding. Similarly, we observed a depletion in our ability to predict DAP binding at sites of high co-occupancy for some factors; however, there was no association between predictive power and degree of co-occupancy for other DAPs, indicating that sequence specificity at sites of high DAP occupancy are factor dependent.
Although some of our observations were similar to what has been observed in vitro, our analysis highlights the value in performing ChIP-seq analysis in primary tissue to characterize tissue-specific gene regulation. The DNA-binding proteins we analyzed show a high degree of promoter-proximal binding near genes uniquely expressed in the liver (Fig. 3C), and these DAP binding events are preferentially enriched in liver-specific eQTL SNPs compared to eQTLs specific to other tissues (Fig. 4B), in agreement with previous analyses of general chromatin marks in primary tissue (Roadmap Epigenomics Consortium et al. 2015). These tissue-specific correlations were diminished in the HepG2 cell line (Fig. 3D), demonstrating that analysis of primary tissue can improve our understanding of in vivo gene regulation and liver pathophysiology. Although cell lines may lack some tissue-specific signals, our data nicely complement HepG2 data specifically in relation to cancer: HepG2-derived DAP binding sites occur more commonly near genes overexpressed in liver tumors, whereas tissue-derived DAP binding sites were more common near genes with decreased expression in tumors (Fig. 6G). Furthermore, tissue-derived DAP binding sites exhibited higher enrichment for somatic mutations compared to flanking regions than that observed in HepG2 cells (Fig. 6H). The complementary nature of our data with existing cell line–based experiments suggests it will facilitate investigations of cis-regulatory element disruption across a variety of liver pathologies.
We have also demonstrated an effective application of these data to prioritize impactful noncoding sequence variations, which we validated by observations of conservation, allele-specific bias in DAP occupancy at sites of heterozygous SNPs (Fig. 5B), and in vitro reporter assays (Fig. 5D,E). Several putative DAP-disruptive eQTL SNPs were associated with relevant phenotypes in liver tissue, including glucose homeostasis, drug metabolism, and circulating lipid levels, and therefore represent a promising resource for future mechanistic follow-up. Despite a limited sample size, our application of this method for prioritizing variants represents an improvement over large agnostic assays that have reported a success rate <5% (Tewhey et al. 2016).
There are important limitations to our study. First, we prioritized the breadth of factors assayed, which constrained us to conducting assays on only two individuals. This limits our ability to construct reasonable estimates of natural variation in DAP occupancy or to identify robust associations between DAP occupancy and donor demographics like age, sex, or ethnicity. In addition, we assayed only a small portion of the known DAPs expressed in humans (Fulton et al. 2009), and repressive factors are particularly underrepresented in our sample. Sampling a larger number of DAPs would likely reveal a more comprehensive picture and uncover additional putatively disruptive regulatory sequence variants. Cell-type heterogeneity is a potential source of noise in our data set and could obfuscate comparisons with independently derived tissue samples. However, we observed no decrease in the number of replicate concordant peaks in our tissue-derived data set compared to HepG2, a high degree of peak overlap across donors, and an enrichment for previously described liver expression programs. Further analysis of homogenous cell types could be complementary to our bulk tissue-derived data set in dissecting the regulatory landscapes of specific cell populations and provide a more detailed understanding of which cell types are most likely affected by trait-associated sequence variation. Previous analyses have also suggested that HepG2 cells exhibit features similar to pediatric hepatoblastoma, potentially confounding our comparisons with adult liver expression and cancer genomic data sets (Pang et al. 2004).
Despite the significant amount of work to be done in fully characterizing the regulatory landscape of the human genome, the application of genomic techniques has shed light on the high level of coordination required for the precise, spatiotemporal control of gene expression. Although painstaking efforts by large consortia have greatly contributed to our understanding of these intricate molecular processes, one notable hurdle that remains is validating functional genomic data generated in cell culture models in tissues. This work will serve as an important resource to the research community and will further facilitate a broad functional genomic investigation of DAP binding in additional human tissues.
Methods
ChIP-seq, WGS, and RNA-seq sample preparation
Liver tissue was obtained from both deceased donors and flash frozen at the time of organ procurement. ChIP-seq was performed in replicate for each DAP using a previously established method (Savic et al. 2013). Antibodies used for ChIP-seq assays are listed in Supplemental Table S16. Binding sites were identified using the MACS peak caller (Zhang et al. 2008). Narrow peaks were defined as 100-bp segments of DNA centered on the peak summit. Replicate BAM files for each factor analyzed in our human tissue donors, except for EGR1, were obtained from previous work in our group in HepG2, which is publicly available at the ENCODE data portal (https://www.encodeproject.org). 10x Chromium, WGS, and phased BAM and VCF files were generated from frozen liver tissue from each donor via the 10x Genomics Longranger pipeline by the HudsonAlpha Genomic Services Lab (https://gsl.hudsonalpha.org/information/10X). RNA was obtained via four independent tissue pulverizations on each liver and extracted using the Norgen Animal Tissue RNA purification kit. RNA-seq libraries were generated using Tn-RNA-seq, a transposase-mediated construction method, as previously described (Gertz et al. 2012b). Sequencing reads were aligned using a previously described pipeline (Alonso et al. 2017). For all analyses, we used hg19 rather than the more recent GRCh38 build in order to facilitate comparative analysis with public data such as GTEx and ENCODE.
Allele-specific binding and expression analysis
Allele bias in DAP occupancy was assessed with ChIP-seq data using the R package “BaalChIP” (DeSantiago et al. 2017). Allele-specific expression was calculated for each expressed heterozygous SNP using the GATK “ASEReadCounter” function according to previously described best practices (Castel et al. 2015).
Gene set–ChIP binding proximity analysis
For a given pathway or gene set, the distribution of distances from the TSS of each gene to the nearest binding site for a given factor was compared to the distribution of TSS to nearest binding site distances for the entire transcriptome as previously described (Savic et al. 2016) and significance was determined using the nonparametric Kolmogorov–Smirnov test.
Support vector machine training
SVMs were trained on replicate-concordant narrow peak sites from the adult liver using a method previously established (Ghandi et al. 2014; Lee 2016). This resulted in 20 SVMs, one for each DAP analyzed. To identify GTEx liver eQTL SNPs likely to modulate DAP binding affinity, we obtained 100 bp of genome sequence centered on each liver eQTL SNP generating two, 100-bp sequence windows containing the reference or the alternate allele. The reference and alternate sequences were scored with the each of the 20 SVMs trained on each DAP analyzed. The reference classifier value was subtracted from the alternate allele to obtain a delta binding score.
The Supplemental Methods includes additional information on sample preparation for ChIP-seq and RNA-seq as well as analysis and SVM training and correlation with public data sets.
Data access
ChIP-seq data from this study have been submitted to the ENCODE data portal (https://www.encodeproject.org) under the sample accession numbers ENCDO882MMZ and ENCDO060 AAA. RNA-seq and WGS data from this study have been submitted to the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE102188 and to the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under the NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/) accession number PRJNA396912, respectively.
Supplementary Material
Acknowledgments
We thank Christopher Partridge for his assistance with plasmid reporter assays and Mike Snyder for sharing liver tissue for analysis. We also thank the HudsonAlpha Genomics Services Laboratory for their contributions to the large-scale DNA sequencing performed for the experiments in this study. This work was supported by the National Institute of Health (NIH) grants U54 HG006998-0 and 5T32GM008361-21, the HudsonAlpha Tie the Ribbons Fund, the UAB CCTS (NIH 1UL1TR001417-01), and funding from the State of Alabama.
Author contributions: D.S., R.M.M., S.J.C., and G.M.C. conceived of the study. D.S. and K.N. prepped samples for RNA-seq and ChIP-seq. R.C.R. and D.S. performed computational analysis. A.A.H. and R.C.R. performed reporter assays. R.C.R., D.S., G.M.C., A.A.H., S.J.C., and R.M.M. contributed to writing the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.222083.117.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Lasseigne BN, Williams K, Nielsen J, Ramaker RC, Hardigan AA, Johnston BE, Roberts BS, Cooper SJ, Marsal S, et al. 2017. aRNApipe: a balanced, efficient and distributed pipeline for processing RNA-seq data in high performance computing environments. Bioinformatics 11: 1727–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. 2012. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem 287: 7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. 2015. Tools and best practices for data processing in allelic expression analysis. Genome Biol 16: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15: 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaForest A, Nagaoka M, Si-Tayeb K, Noto FK, Konopka G, Battle MA, Duncan SA. 2011. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development 138: 4143–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Alpern D, Gardeux V. 2016. The genetics of transcription factor DNA binding variation. Cell 166: 538–554. [DOI] [PubMed] [Google Scholar]
- DeSantiago I, Liu W, Yuan K, O'Rielly M, Chilamakuri C, Ponder B, Meyer K, Markowetz F. 2017. BaalChIP: Bayesian analysis of allele-specific transcription factor binding in cancer genomes. Genome Biol 18: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan N, Wu W, Morrissey CS, Chen KB, Stonestrom A, Long M, Keller CA, Cheng Y, Jain D, Visel A, et al. 2015. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than histone modifications or chromatin accessibility. Epigenetics Chromatin 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. 2013. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93: 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. 2006. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer 5: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Desjardins CA, Cooper O, Kontor A, Nocco SE, Naya FJ. 2015. EGR1 functions as a potent repressor of MEF2 transcriptional activity. PLoS One 10: e0127641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletez-Brant C, Lee D, McCallion AS, Beer MA. 2013. kmer-SVM: a web server for identifying predictive regulatory sequence features in genomic data sets. Nucleic Acids Res 41: W544–W556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. 2009. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol 10: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvagni F, Capo S, Oliviero S. 2001. Sp1 and Sp3 physically interact and co-operate with GABP for the activation of the utrophin promoter. J Mol Biol 306: 985–996. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Reddy TE, Varley KE, Garabedian MJ, Myers RM. 2012a. Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome Res 22: 2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Varley KE, Davis NS, Baas BJ, Goryshin IY, Vaidyanathan R, Kuersten S, Myers RM. 2012b. Transposase mediated construction of RNA-seq libraries. Genome Res 22: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. 2013. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell 52: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Lee D, Mohammad-Noori M, Beer MA. 2014. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput Biol 10: e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 12: 393–406.21502959 [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1503. [DOI] [PubMed] [Google Scholar]
- Kaiser VB, Taylor MS, Semple CA. 2016. Mutational biases drive elevated rates of substitution at regulatory sites across cancer types. PLoS Genet 12: e1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlebach G, Shamir R. 2008. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol 9: 770–780. [DOI] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. 2016. Role of non-coding sequence variants in cancer. Nat Rev Genet 17: 93–108. [DOI] [PubMed] [Google Scholar]
- Landt S, Marinov G. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. 2016. LS-GKM: a new gkm-SVM for large-scale datasets. Bioinformatics 32: 2196–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS, Beer MA. 2015. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 47: 955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chanrion M, Sawey E, Wang T, Chow E, Tward A, Su Y, Xue W, Lucito R, Zender L, et al. 2015. Reciprocal interaction of Wnt and RXR-α pathways in hepatocyte development and hepatocellular carcinoma. PLoS One 10: e0118480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield LM, Klinge CM. 2012. Multiple roles of COUP-TFII in cancer initiation and progression. J Mol Endocrinol 49: R135–R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Jimenez CP, Kyrmizi I, Cardot P, Gonzalez FJ, Talianidis I. 2010. Hepatocyte nuclear factor 4α coordinates a transcription factor network regulating hepatic fatty acid metabolism. Mol Cell Biol 30: 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. 2013. Identification of genetic variants that affect histone modifications in human cells. Science 342: 747–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17: 17–43. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Pepke S, Jansen C, Marinov GK, Ernst J, Kellis M, Hardison RC, Myers RM, Wold BJ. 2013. Integrating and mining the chromatin landscape of cell-type specificity using self-organizing maps. Genome Res 23: 2136–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Yoneda Y. 2001. Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin β. Mol Cell Biol 21: 2779–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. 2012. Predicting cell-type–specific gene expression from regions of open chromatin. Genome Res 22: 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X, et al. 2010. Hepatocyte nuclear factor 4α suppresses the development of hepatocellular carcinoma. Cancer Res 70: 7640–7651. [DOI] [PubMed] [Google Scholar]
- Pang RT, Poon TC, Wong N, Lai PB, Wong NL, Chan CM, Yu JW, Chan AT, Sung JJ. 2004. Comparison of protein expression patterns between hepatocellular carcinoma cell lines and a hepatoblastoma cell line. Clin Proteomics 1: 313–332. [Google Scholar]
- Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA, et al. 2015. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. 2012a. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol 32: 3756–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Gertz J, Pauli F, Kucera KS, Varley KE, Newberry KM, Marinov GK, Mortazavi A, Williams BA, Song L, et al. 2012b. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res 22: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. 2004. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein–protein interactions. Blood Cells, Mol Dis 32: 143–154. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Ernberg I. 2005. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). Proc Natl Acad Sci 102: 2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic D, Gertz J, Jain P, Cooper GM, Myers RM. 2013. Mapping genome-wide transcription factor binding sites in frozen tissues. Epigenetics Chromatin 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, Mackiewicz M, Mendenhall EM, Myers RM. 2015a. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res 25: 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic D, Roberts BS, Carleton JB, Partridge EC, White MA, Cohen BA, Cooper GM, Gertz J, Myers RM. 2015b. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/enhancer-binding protein beta binding sites. Genome Res 25: 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic D, Ramaker RC, Roberts BS, Dean EC, Burwell TC, Meadows SK, Cooper SJ, Garabedian MJ, Gertz J, Myers RM, et al. 2016. Distinct gene regulatory programs define the inhibitory effects of liver X receptors and PPARG on cancer cell proliferation. Genome Med 8: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Sur I, Taipale J. 2016. The role of enhancers in cancer. Nat Rev Cancer 16: 483–493. [DOI] [PubMed] [Google Scholar]
- Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, Goldring MB. 2003. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1β. J Biol Chem 278: 17688–17700. [DOI] [PubMed] [Google Scholar]
- Tehranchi AK, Myrthil M, Martin T, Hie BL, Golan D, Fraser HB. 2016. Pooled ChIP-seq links variation in transcription factor binding to complex disease risk. Cell 165: 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich T. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK, Andersen KG, Mikkelsen TS, Lander ES, Schaffner SF, et al. 2016. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhao Y, Xia N, Zhang W, Tang Z, Wang C, Zhu X, Cui S. 2015. KPNβ1 promotes palmitate-induced insulin resistance via NF-κB signaling in hepatocytes. J Physiol Biochem 71: 763–772. [DOI] [PubMed] [Google Scholar]
- Xie D, Boyle AP, Wu L, Zhai J, Kawli T, Snyder M. 2013. Dynamic trans-acting factor colocalization in human cells. Cell 155: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. 2013. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154: 801–813. [DOI] [PubMed] [Google Scholar]
- Yu S, Cui K, Jothi R, Zhao D, Jing X, Zhao K, Xue H. 2016. GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood 117: 2166–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang G, Qian J. 2016. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet 17: 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.