Abstract
In eukaryotic cells RNAs at various maturation and processing levels are distributed across cellular compartments. The standard approach to determine transcript abundance and identity in vivo is RNA sequencing (RNA-seq). RNA-seq relies on RNA isolation from whole cell lysates and thus mainly captures fully-processed, stable and more abundant cytoplasmic RNAs over nascent, unstable and nuclear RNAs. Here, we provide a step-by-step protocol for subcellular RNA-seq (subRNA-seq). subRNA-seq allows the quantitative measurement of RNA polymerase II-generated RNAs from the chromatin, nucleoplasm and cytoplasm of mammalian cells. This approach relies on cell fractionation prior to RNA isolation and sequencing library preparation. High-throughput sequencing of the subcellular RNAs can then be used to reveal the identity, abundance and subcellular distribution of transcripts and thus provides insights into RNA processing and maturation. Deep sequencing of the chromatin-associated RNAs further offers the opportunity to study nascent RNAs. Subcellular RNA-seq libraries are obtained within 5 days.
Keywords: Cell fractionation, subcellular RNA-seq, nascent RNA, RNA polymerase II (Pol II), transcription, next-generation sequencing, RNA processing
INTRODUCTION
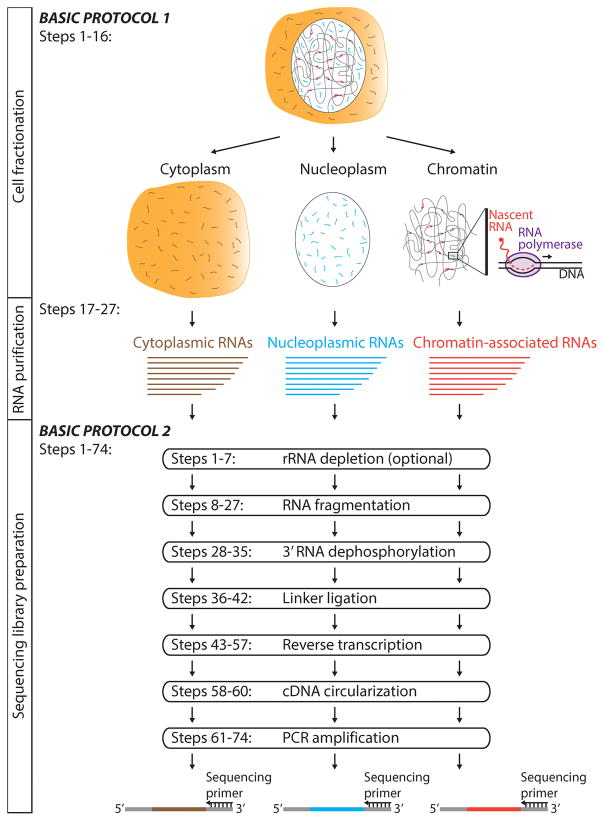
Eukaryotic genomes are pervasively transcribed by RNA polymerase II (Pol II) giving rise to all messenger RNAs and a large set of non-coding RNAs (Djebali et al., 2013; Jensen et al., 2013; Pelechano and Steinmetz, 2013). Nascent RNA is synthesized in a chromatin environment by stable Pol II transcription complexes (Jonkers and Lis, 2015; Venkatesh and Workman, 2015). At the poly-adenylation (pA) site the nascent transcript is cleaved and poly-adenylated. The RNA that has been properly processed, as controlled by RNA surveillance mechanisms, is released into the nucleoplasm and further processed before it is exported into the cytoplasm (Kilchert et al., 2016). The RNA export is tightly regulated in vivo and involves many export factors (Oeffinger and Zenklusen, 2012; Meinel and Sträßer, 2015; Wickramasinghe and Laskey, 2015). The fully processed messenger RNA (mRNA) serves as a blueprint for protein synthesis in the cytoplasm. Thus, Pol II transcripts at different processing and maturation levels are distributed between distinct cellular compartments, mainly between chromatin (nascent RNA), nucleoplasm (processed and unprocessed RNA) and cytoplasm (mature RNA) (Figure 1).
Figure 1.
Schematic overview of the key steps of the subRNA-seq approach.
The standard approach to globally measure transcript abundance and identity is RNA preparation coupled with high-throughput sequencing (RNA-seq) (Wang et al., 2009). The standard RNA-seq method relies on the preparation of whole cell lysates prior to RNA isolation. Therefore, the information about the subcellular location of a given RNA is lost. Furthermore, the generation of whole cell extracts also results in sequencing libraries that are dominated by fully-processed stable RNAs that mainly occur in the cytoplasm. These cytoplasmic RNAs usually complicate the analysis of nascent and unstable transcripts.
To overcome these limitations new variants of the original RNA-seq method have been recently developed by us and others (Mayer et al., 2015; Bhatt et al., 2012; Pandya-Jones and Black, 2009; Tilgner et al., 2012; Zaghlool et al., 2013; Ferrari et al., 2013; Weber et al., 2014; Conrad et al., 2014; Mondal et al., 2010). A communality among these approaches is that they rely on cell and chromatin fractionation prior to RNA purification and sequencing library preparation.
Here we provide a step-by-step protocol for the genome-wide measurement of transcript abundance and identity in different cellular compartments –chromatin, nucleoplasm, cytoplasm– by subcellular RNA-seq (subRNA-seq). This approach combines effective cell fractionation with robust and efficient sequencing library preparation to reveal the relative abundance and the identity of transcripts in different cellular compartments. subRNA-seq reveals the full spectrum of Pol II synthesized RNAs and at different processing levels, ranging from unprocessed nascent RNAs to fully processed cytoplasmic RNAs. The approach has been successfully applied to HeLa S3 and HEK293T and will be readily applicable to other mammalian cell types (Mayer et al., 2015). Thus, subRNA-seq represents a powerful genomic tool for the analysis of RNA processing and also provides an opportunity to study nascent RNAs in a broad range of mammalian cell types.
BASIC PROTOCOL 1. CELL FRACTIONATION & RNA ISOLATION
The cell fractionation approach described here is based on protocols that have been developed by us and others (Wuarin and Schibler, 1994; Pandya-Jones and Black, 2009; Bhatt et al., 2012; Mayer et al., 2015). It is most similar to the cell fractionation protocol that we have established for human native elongating transcript sequencing (NET-seq) (Mayer and Churchman, 2016) (Figure 1). Cells are lysed and the cytoplasm is separated from nuclei by centrifugation through a sucrose cushion. The nuclei pellet is washed to remove cytoplasmic remnants prior to chromatin fractionation. The chromatin is separated from the nucleoplasm in the presence of Urea, the nonionic detergent Nonidet P-40 (NP-40) and NaCl as originally described by the Schibler lab (Wuarin and Schibler, 1994). Next, the RNA is prepared in parallel from the chromatin, nucleoplasmic and cytoplasmic fractions. Remaining DNA is removed by a standardized DNase I digest.
Cell fractionation is performed in the presence of the Pol II transcription inhibitor α-amanitin (Lindell et al., 1970; Brueckner and Cramer, 2008) and RNase inhibitors to avoid run-off transcription and RNA degradation during sample processing, respectively.
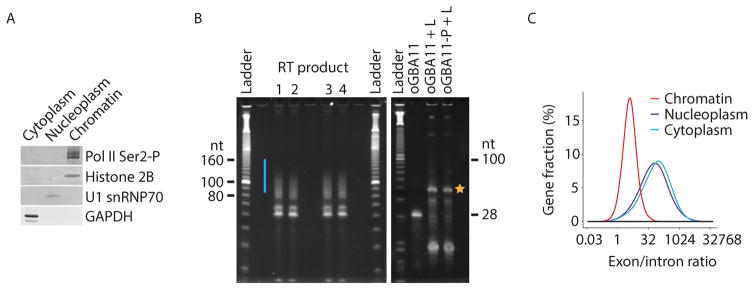
We recommend to monitor the success of the cell fractionation experiment by Western blot analysis (Figure 2A) using antibodies against subcellular markers. This is especially true, when the cell fractionation approach is applied to new cell types.
Figure 2. Key quality controls and expected results from subRNA-seq experiments.
(A) Representative Western blot of subcellular fractions. The different cellular fractions obtained from HeLa S3 cells were probed against transcribing Pol II (Pol II Ser2-P; 3E8 antibody) the chromatin marker Histone 2B (FL-126 antibody), the nucleoplasmic marker U1 snRNP70 (C-18 antibody) and the cytoplasmic marker GAPDH (6C5 antibody). The volumes of the different cellular fractions have been adjusted so that the Western blot signals can be compared between the fractions. This figure was modified from (Mayer et al., 2015).
(B) Representative gels showing the product of the reverse transcription (RT) of the samples (left panel) and of the oGAB11 controls (right panel). The region that is excised from the gel is indicated by a blue bar or yellow star for the samples and the oGAB11 controls, respectively.
(C) Distributions of the size-normalized ratio of subRNA-seq reads that map to exons versus introns for each gene. subRNA-seq libraries were generated from HeLa S3 cells. This figure was adapted from (Mayer et al., 2015).
Materials
Cultured mammalian cells: 1x 107 cells per experiment
1x PBS (Life Technologies, cat. no. 10010-023)
α-amanitin (Sigma, cat. no. A2263)
DTT (0.1 M; part of the SuperScript III First-Strand Synthesis System, Life Technologies, cat. no. 18080-051)
NP-40 (Life Technologies, cat. no. 28324)
Triton X-100 (Sigma, cat. no. T9284)
Tween 20 (Sigma, cat. no. 274348)
Sucrose (Sigma, cat. no. S0389)
Glycerol (Sigma, cat. no. G5516)
Urea (Sigma, cat. no. U6504)
Sodium acetate, RNase-free (3 M; Life Technologies, cat. no. AM9740)
NaCl, RNase-free (5 M; Life Technologies, cat. no. AM9760G)
EDTA, RNase-free (0.5 M; Life Technologies, cat. no. AM9260G)
Tris-HCl, RNase-free (1 M, pH 7.0; Life Technologies, cat. no. AM9850G)
Tris-HCl, RNase-free (1 M, pH 8.0; Life Technologies, cat. no. AM9855G)
HEPES, RNase-free (1 M, pH 7.5; Teknova, cat. no. H1035)
NaOH (Pellets; EMD Millipore, cat. no. 106498)
HCl (37% (wt/vol); EMD Millipore, cat. no. HX0607)
SUPERase.In (20 U/μl; Life Technologies, cat. no. AM2696)
Protease inhibitor mix cOmplete, EDTA-free (Roche, cat. no. 11873580001)
GlycoBlue (15 mg/ml; Life Technologies, cat. no. AM9515)
TRI reagent (MRC, cat. no. TR 118)
Chloroform (Sigma, cat. no. 288306)
RLT buffer (Qiagen, cat. no. 79216)
100% (vol/vol) ethanol (VWR, cat. no. V1016)
RNeasy Mini Kit (Qiagen, 74104)
RNase-Free DNase Set (Qiagen, cat. no. 79254)
RNase/DNase-free H2O (Life Technologies, cat. no. 10977-015)
Pol II Ser-2P antibody (3E8; Active Motif, cat. no. 61083)
Histone 2B antibody (FL-126; Santa Cruz Biotechnology, cat. no. sc-10808)
U1 snRNP70 antibody(C-18; Santa Cruz Biotechnology, cat. no. sc-9571)
GAPDH antibody (6C5; Life Technologies, cat. no. AM4300)
The antibodies listed above are optional.
CAUTION: α-amanitin is toxic. Handle solutions containing α-amanitin with care and dispose waste according to the institutional regulations.
CAUTION: DTT is toxic and corrosive. DTT causes irritations. Solutions containing DTT should be used with care.
CAUTION: NP-40 causes irritations. Solutions containing NP-40 should be used with care.
CAUTION: Triton X-100 is harmful and causes irritations. Triton X-100 is hazardous to the environment. Solutions containing Triton X-100 should be used with care.
CAUTION: EDTA causes irritations. Solutions containing EDTA should be used with care.
CAUTION: The TRI reagent contains phenol and guanidine thiocyanate. Phenol is toxic and guanidine thiocyanate causes irritations. Handle the TRI reagent with care and dispose according to institutional regulations.
CAUTION: Chloroform is volatile and toxic. Chloroform causes irritations. Handle solutions containing chloroform with care and dispose chloroform waste according to the institutional regulations.
CAUTION: Use the following components of the RNeasy mini kit with care. The RWT buffer is corrosive and causes irritations. The Qiazol Lysis Reagent is toxic and corrosive. Use personal protective equipment when handling this kit and dispose waste according to the institutional regulations.
Cell fractionation
All experimental steps of the cell fractionation are performed on ice or at 4°C. Pre-cooled buffers are used.
-
1
Use 1x 107 cells as an input per experiment.
The amount of input refers to HeLa S3 and HEK293T cells. Although we expect that the number of cells which are required for a successful subRNA-seq experiment should be similar for other mammalian cell types, in some cases protocol adjustments might be necessary as described in the COMMENTARY section. We recommend to always determine the number of cells by cell counting. -
2
Wash cells 2x with 10 ml 1x PBS.
-
3
Scrape cells into 9 ml 1x PBS.
This experimental step refers to cells that grow adhesively or semi-adhesively. -
4
Centrifuge for 2 min at 500 g at 4°C. Discard supernatant.
-
5
Cut pipette tips and resuspend cell pellets in 200 μl Cytoplasmic lysis buffer.
-
6
Incubate on ice for 5 min.
-
7
Cut tips and layer the cell lysate onto 500 μl Sucrose buffer.
-
8
Centrifuge for 10 min at 16,000 g at 4°C.
-
9
Transfer the supernatant to a new tube and store on ice until further processed (see step 23).
The supernatant represents the cytoplasmic fraction. -
10
Wash nuclei pellet with 800 μl Nuclei wash buffer.
-
11
Spin for 1 min at 1,150 g at 4°C. Discard supernatant.
-
12
Cut tips and gently resuspend nuclei in 200 μl Glycerol buffer.
-
13
Add 200 μl Nuclei lysis buffer.
-
14
Mix by pulsed vortexing and incubate on ice for 2 min.
-
15
Centrifuge 2 min at 18,500 g at 4°C.
-
16
Transfer the supernatant to a new tube and store on ice until further processed (see step 23) and also keep the pellet.
The supernatant and pellet represent the nucleoplasmic and chromatin fraction, respectively.
Isolation of chromatin-associated, nucleoplasmic and cytoplasmic RNAs
Chromatin RNA
-
17
Add 50 μl pre-cooled 1x PBS to the chromatin pellet (from step 16) and resuspend by pipetting.
-
18
Add 500 μl TRI reagent and mix resuspend the pellet by vortexing.
-
19
Add 100 μl chloroform, vortex and incubate at RT for 5 min.
-
20
Centrifuge for 15 min at 16,000 g at 4°C.
-
21
Transfer the upper aqueous layer to a new tube, add 3.5x sample volumes of RLT buffer and mix.
-
22
Add 2.5x of the initial sample volume (before the RLT buffer was added) ice-cold 100% (vol/vol) ethanol and mix. Proceed with step 25.
Cytoplasmic and nucleoplasmic RNA
-
23
Add 3.5x sample volumes RLT buffer and mix.
-
24
Add 2.5x of the initial sample volume (before the RLT buffer was added) ice-cold 100% (vol/vol) ethanol and mix. Proceed with step 25.
Chromatin, cytoplasmic and nucleoplasmic RNA
-
25
Follow clean up protocol of the RNeasy Kit (Qiagen) according to manufacturer’s instructions.
-
26
Perform an on-column DNase I digest (Qiagen) according to manufacturer’s instructions.
-
27
Elute in 50 μl RNase/DNase-free H2O.
Proceed with the sequencing library preparation immediately or store the RNA at −80°C.The RNA concentration as determined by NanoDrop spectrophotometer measurement for the cytoplasmic, nucleoplasmic and chromatin fractions is typically 3000–4000, 600–700 and 400–500 ng/μl, respectively.
BASIC PROTOCOL 2. SEQUENCING LIBRARY PREPARATION
The second part of the subRNA-seq approach exploits an efficient sequencing library preparation method. The library preparation is based on protocols that we developed for the human NET-seq approach (Mayer et al., 2015; Mayer and Churchman, 2016) and is also related to library generations used for yeast NET-seq (Churchman and Weissman, 2012a; 2012b) and ribosome profiling (Ingolia et al., 2012).
Here, the chromatin, nucleoplasmic and cytoplasmic RNAs obtained in protocol 1 steps 1 to 27 are converted into DNA sequencing libraries. This is accomplished by the following major steps: (1) ribosomal RNAs (rRNAs) are depleted from the subcellular RNAs using a commercially available removal kit. (2) RNA is fragmented by partial alkaline hydrolysis. (3) 3′-phosphate groups are removed from the fragmented RNA. (4) A barcoded DNA-linker is ligated to the 3′-ends of the RNA. (5) RNA is converted into cDNA, (6) circularized and (7) amplified using a limited number of PCR cycles (Figure 1).
Each of these key steps was optimized to work at a high efficiency and the success is monitored by quality controls. In case the library passes all quality checks, it is subjected to an Illumina high-throughput sequencing platform such as MiSeq, HighSeq and NextSeq.
Although we describe a NET-seq-based library preparation method here, we expect that our cell fractionation and RNA isolation approach (BASIC PROTOCOL 1) can also be combined with alternative library preparation protocols to obtain sequencing libraries.
Materials
RNase/DNase-free H2O (Life Technologies, cat. no. 10977-015)
Ribo-Zero rRNA Removal Kit (Epicentre, cat. no. MRZH116)
80% (vol/vol) ethanol (VWR, cat. no. V1016)
Sodium carbonate anhydrous (VWR, cat. no. M138)
Sodium bicarbonate (VWR, cat. no. 3509)
EDTA (0.5 M; Life Technologies, cat. no. AM9260G)
Isopropanol (Sigma, cat. no. 278475)
RNA control ladder (0.1–2 kb; Life Technologies, cat. no. 15623-100)
2x TBU denaturing loading buffer (Life Technologies, cat. no. LC6876)
TBE-urea gels, 15% (wt/vol) (Life Technologies, cat. no. EC68852BOX)
TBE buffer, 10x (Life Technologies, cat. no. 15581-044)
Orange G (Sigma, cat. no. O3756)
SYBR Gold Nucleic Acid Gel Stain (10,000x concentrate; Life Technologies, cat. no. S-11494)
PEG8000 (part of T4 RNA Ligase 2, truncated, NEB, cat. no. M0242S)
DMSO (Sigma, cat. no. D8418)
T4 RNA Ligase Buffer, 10x (part of T4 RNA Ligase 2, truncated, NEB, cat. no. M0242S)
T4 RNA Ligase 2, truncated (NEB, cat. no. M0242S)
GlycoBlue (15 mg/ml; Life Technologies, cat. no. AM9515)
Sodium acetate, RNase-free (3 M; Life Technologies, cat. no. AM9740)
T4 PNK Buffer, 10x (part of T4 Polynucleotide Kinase, NEB, cat. no. M0201S)
T4 Polynucleotide Kinase (10,000 U/ml; NEB, cat. no. M0201S)
DTT (0.1 M; part of the SuperScript III First-Strand Synthesis System, Life Technologies, cat. no. 18080-051)
SUPERase.In (20 U/μl; Life Technologies, cat. no. AM2696)
5x First-Strand Buffer (part of SuperScript III First-Strand Synthesis System, Life Technologies, cat. no. 18080-051)
dNTP mix (10 mM; Life Technologies, cat. no. 18427-013)
SuperScript III First-Strand Synthesis System (Life Technologies, cat. no. 18080-051)
TBE-urea gels, 10% (wt/vol) (Life Technologies, cat. no. EC68752BOX)
CircLigase Reaction Buffer, 10x (part of CircLigase ssDNA Ligase, Epicentre, cat. no. CL4111K)
ATP, 1 mM (part of CircLigase ssDNA Ligase, Epicentre, cat. no. CL4111K)
MnCl2, 50 mM (part of CircLigase ssDNA Ligase, Epicentre, cat. no. CL4111K)
CircLigase ssDNA Ligase (100 U/μl; Epicentre, cat. no. CL4111K)
Phusion HF Buffer, 5x (part of Phusion High-Fidelity DNA Polymerase, NEB, cat. no. M0530S)
Phusion DNA Polymerase (2,000 U/ml; NEB, cat. no. M0530S)
DNA control ladder (10 bp; Life Technologies, cat. no. 10821-015)
TBE gels, 8% (wt/vol) (Life Technologies, cat. no. EC62152BOX)
Qubit dsDNA HS Assay Kit (Life Technologies, cat. no. Q32851)
High Sensitivity DNA Analysis Kit (Agilent Technologies, cat. no. 5067-4626)
Scalpels (Electron Microscopy Sciences, cat. no. 72042-11)
20G Needle (BD, cat. no. 305175)
Microfuge tube filter: Costar Spin-X centrifuge tube filters (Sigma, cat. no. CLS8162-96EA)
CAUTION: Sodium carbonate causes irritations. Solutions containing sodium carbonate should be used with care.
CAUTION: EDTA causes irritations. Solutions containing EDTA should be used with care.
CAUTION: NaOH-containing solutions are corrosive. Handle solutions containing NaOH with care and dispose waste according to the institutional regulations.
CAUTION: HCl-containing solutions are corrosive and cause irritations. Handle solutions containing HCl with care and dispose waste according to the institutional regulations
CAUTION: Protease inhibitor mix cOmplete causes irritations. Solutions containing protease inhibitors should be used with care.
CAUTION: Isopropanol is highly flammable and volatile. Isopropanol causes irritations. Isopropanol should be used with care.
CAUTION: Ethanol is highly flammable and volatile. Ethanol causes irritations. Ethanol should be used with care.
CAUTION: Chloroform is volatile and toxic. Chloroform causes irritations. Handle solutions containing chloroform with care and dispose chloroform waste according to the institutional regulations.
CAUTION: SYBR Gold Nucleic Acid Gel Stain is flammable. Nucleic acid stains are usually mutagenic. Use personal protective equipment when handling nucleic acid gel stains and dispose waste according to the institutional regulations.
CAUTION: MnCl2 is toxic and hazardous to the environment. Handle solutions containing MnCl2 with care and dispose waste according to the institutional regulations.
CAUTION: Handle sharps (such as scalpels and needles) with care. Dispose sharps according to the institutional regulations.
rRNA depletion
In order to increase the fraction of informational sequencing reads that originate from Pol II-synthesized transcripts, ribosomal RNAs are depleted. The rRNA depletion is optional.
-
1
Use 20 μg of RNA as input per depletion.
-
2
Apply the Ribo-Zero rRNA Removal Kit (Epicentre) and follow the manufacturer’s instructions.
-
3
Following the RNA precipitation (as described in the manufacturer’s instructions), centrifuge 30 min at 20,000 g at 4°C. Discard supernatant.
-
4
Wash RNA pellet with 750 μl pre-cooled 80% (vol/vol) ethanol, quick spin and discard supernatant.
-
5
Repeat ethanol wash (step 4) one more time.
-
6
Air-dry pellet at room temperature for approximately 10 min.
-
7
Resuspend the RNA pellet in 10 μl RNase/DNase-free H2O.
Proceed with RNA fragmentation immediately or store at −80°C.
RNA Fragmentation
RNA fragmentation is performed by partial alkaline hydrolysis as described in detail in the human and yeast NET-seq protocols (Mayer and Churchman, 2016; Churchman and Weissman, 2012b). The fragmentation time needs to be adjusted for new batches of alkaline fragmentation solution. The length of the majority of the fragmented RNA should be in the range that is excised from the polyacrylamide gel as described below.
-
8
Denature ≥ 3 μg of rRNA-depleted RNA for 2 min at 80°C and quick spin.
-
9
Prepare the fragmentation mix as follows:
Component Amount per reaction (μl) Final PEG8000 (50%) 8.0 20% (vol/vol) DMSO 2.0 10% (vol/vol) T4 RNA Ligase Buffer (10x) 2.0 1x EDTA (0.5 M) 0.7 17.5 mM RNA (1 μg/μl) 1.0 1 μg RNase/DNase-free H2O 6.3 Prepare three fragmentation mixes per sample. -
10
Add 20 μl of 2x Alkaline fragmentation solution to each sample and mix.
-
11
Fragment RNA for the adjusted time at 95°C in a thermal cycler. Place immediately on ice.
The fragmentation time needs to be adjusted for each new batch of alkaline fragmentation solution. Fragmentation times between 10 to 40 min should be tested. The size of most RNA fragments should be between 30 and 100 nt. -
12
Add sample to 562 μl RNA precipitation solution and mix. Add 750 μl isopropanol and mix. Precipitate the fragmented RNA for ≥1 h at −80°C.
-
13
Centrifuge at 20,000 g for 30 min at 4°C.
-
14
Discard supernatant and wash RNA pellet with 750 μl 80% (vol/vol) ice-cold ethanol.
-
15
Air-dry RNA pellet for 10 min at room temperature
-
16
Resuspend pellet in 10 μl RNase/DNase-free H2O.
-
17
Prepare the RNA control ladder and the oGAB11 control (Table 1). Typically, add 9 μl of RNase/DNase-free H2O to 1 μl RNA control ladder and the oGAB11 control. Add 10 μl of 2x TBU denaturing loading buffer to the RNA sample, the RNA control ladder and the oGAB11 control.
-
18
Denature the RNA sample, the RNA control ladder and the oGAB11 control for 2 min at 80°C.
-
19
Pre-run a 15% TBE-urea polyacrylamide gel for 15 min at 200 V according to the manufacturer’s instructions.
-
20
Load the fragmented RNA sample and the RNA control ladder and run the 15% TBE-urea gel for 65 min at 200 V in 1x TBE.
-
21
Stain the gel for 5 min at room temperature in the Gel staining solution and visualize the fragmented RNA, the RNA control ladder and the oGAB11 control under blue or UV light. Excise the fragmented RNA between 30 and 100 nt.
TABLE 1.
DNA and RNA oligos required for the preparation of subcellular RNA-seq libraries
| DNA oligos | |
| Linker-1a,b,c | /5rApp/CTGTAGGCACCATCAAT/3ddC |
| oLSC007 (RT primer)d,e | /5Phos/ATCTCGTATGCCGTCTTCTGCTTG/iSp18/CACTCA/iSp18/TCCGACGATCATTGATGGTGCCTACAG |
| oNTI231 (reverse primer) | CAAGCAGAAGACGGCATACGA |
| oLSC006 (sequencing primer) | TCCGACGATCATTGATGGTGCCTACAG |
| RNA oligos | |
| oGAB11 (control oligo) | agucacuuagcgauguacacugacugug |
| oGAB11-Pf | agucacuuagcgauguacacugacugug/3Phos/ |
5rApp: 5′-Riboadenylate
3ddC: 3′-Dideoxycytidine
Commercially available from Integrated DNA Technologies
5Phos: 5′-Phosphate
iSp18: internal 18-atom hexa-ethylenglycol spacer
3Phos: 3′-Phosphate
Rapid gel extraction of the fragmented RNA
The rapid gel extraction approach is based on a protocol originally developed by Ingolia et al. (Nat. Protocols, 2012) and is described in detail by Mayer & Churchman (Nat. Protocols, 2016).
-
22
Pierce a 0.5 ml RNase/DNase-free non-stick tube with a 20G needle and place it inside a 1.5 ml RNase/DNase-free non-stick tube. Transfer the gel slice into the pierced 0.5 ml RNase/DNase-free non-stick tube. Centrifuge for 3 min at 20,000 g at room temperature.
-
23
Add 200 μl RNase/DNase-free H2O and incubate for 10 min at 70°C. Vortex for 30 sec.
-
24
Transfer the gel slurry to a microcentrifuge tube filter. Centrifuge for 3 min at 20,000 g at room temperature.
-
25
Combine the three eluates per sample (~600 μl) and transfer to a new 1.5 ml RNase/DNase-free non-stick tube. Add 2 μl GlycoBlue (15 mg/ml) and 50 μl of 3 M sodium acetate (pH 5.5). Mix well. Add 750 μl isopropanol and mix. Incubate for ≥1 h at −80°C.
-
26
Recover the RNA as described in steps 13 to 15.
-
27
Resuspend the RNA pellet in 22.5 μl RNase/DNase-free H2O.
The sample can be stored up to 3 months at −80°C.
3′ RNA dephosphorylation
RNA fragmentation by partial alkaline hydrolysis not only results in RNA molecules with a 3′-OH but also generates RNAs that contain a 3′-phosphate. The 3′-phosphate would prevent a successful DNA linker ligation and therefore need to be removed before ligation. This is accomplished by enzymatic dephosphorylation using the T4 Polynucleotide Kinase (PNK). The T4 PNK catalyzes 3′ RNA dephosphorylation in the absence of ATP. The dephosphorylation procedure described here is based on a protocol published by the Weissman lab (Ingolia et al., 2013).
The success of the enzymatic 3′ RNA dephosphorylation can be monitored by using the 3′-phosphorylated RNA control oligo oGAB11-P (Table 1, Figure 2B, right panel).
-
28
Prepare the T4 PNK reaction mix as follows:
Component Amount for 2 reactions (μl) Final T4 PNK Buffer, 10x 10 2x SUPERase.In (20 U/μl) 2 40 U RNase/DNase-free H2O 37.5 -
29
Denature 22.5 μl of fragmented RNA (from step 27) and 22.5 μl (0.1 μg) of oGAB11-P control oligo for 2 min at 80°C. Cool the denatured RNA on ice for 3 min.
-
30
Add 25 μl T4 PNK reaction mix (from step 28) to each tube and mix. Add 2.5 μl T4 PNK (10 U/μl) and carefully mix.
-
31
Incubate the samples for 1 h at 37°C followed by 10 min at 75°C to inactivate the T4 PNK enzyme.
-
32
Add 450 μl RNase/DNase-free H2O, 56 μl of 3M sodium acetate, 2 μl of 15 mg/ml GlycoBlue and mix. Add 600 μl isopropanol and mix.
-
33
Precipitate the 3′ dephosphorylated RNA and the dephosphorylated oGAB11 control for ≥1 h at −20°C.
-
34
Recover the RNA and the dephosphorylated oGAB11 control as described in steps 13 to 15.
-
35
Resuspend pellets in 6 μl pre-cooled RNase/DNase-free H2O.
Ligation of DNA linker-1
-
36
Denature dephosphorylated RNA, the dephosphorylated oGAB11 control (from step 35) and the oGAB11 control (1 μg in 6 μl) (Table 1) for 2 min at 80°C.
-
37
Prepare the Linker ligation mix for the RNA sample, the dephosphorylated oGAB11 control and the oGAB11 control as follows:
Component Amount per reaction (μl) Final PEG8000 (50% vol/vol) 8.0 20% (vol/vol) DMSO 2.0 10% (vol/vol) T4 RNA Ligase Buffer (10x) 2.0 1x DNA linker-1 (1 μg) 1.0 RNA sample/oGAB11 controls 6.0 Truncated T4 RNA Ligase 2 1.0 200 U -
38
Perform the ligation reaction for 3 h at 37°C. Add 0.7 μl of 0.5 M EDTA and mix to stop the ligation.
-
39
Add 562 μl RNA precipitation solution and mix. Add 750 μl isopropanol and mix.
-
40
Precipitate the linker-ligated RNA and the oGAB11 controls for ≥ 1 h at −20°C.
-
41
Recover the RNA and the oGAB11 controls as described in steps 13 to 15.
-
42
Resuspend pellets in 10 μl RNase/DNase-free H2O.
Reverse transcription
-
43
Prepare the Reverse transcription mix as follows:
Component Amount per reaction (μl) Final 5x First-Strand Buffer 3.3 1x dNTPs (10 mM) 0.8 0.5 mM Reverse primer oLSC007 (10 μM) 0.5 0.3 μM -
44
Add 10 μl of linker-ligated RNA or of the linker-ligated oGAB11 controls (step 42), mix and incubate for 2 min at 80°C. Put the sample on ice for 3 min.
-
45
Add 1.3 μl SUPERase.IN/DTT mix to each tube and mix. Add 0.8 μl of SuperScript III (200 U/μl) and mix.
-
46
Incubate the reverse transcription mixture for 30 min at 48°C. Add 1.8 μl of 1N NaOH, mix and incubate for 20 min at 98°C. Neutralize the reaction by adding 1.8 μl of 1N HCl, mix and put on ice.
-
47
Add 20 μl of 2x TBU denaturing sample to each tube and mix.
-
48
Also prepare the DNA control ladder by adding 1.0 μl of DNA control ladder to 9 μl RNase/DNase-free H2O. Add 10 μl of 2x TBU denaturing sample buffer and mix.
-
49
Denature the cDNA samples (including the oGAB11 controls) and the DNA control ladder for 3 min at 95°C. Put the samples on ice for 3 min.
-
50
Pre-run a 10% (wt/vol) polyacrylamide TBE-urea gel at 200 V for 15 min in 1x TBE.
-
51
Separate the cDNA samples and the DNA control ladder by PAGE at 200 V for 65 min.
-
52
Stain the gel in 50 ml of Gel staining solution for 5 min at room temperature.
The signal intensity of the dephosphorylated oGAB11 control and of the oGAB11 control should be similar, indicating a high dephosphorylation efficiency (Figure 2B, right panel). -
53
Excise the cDNA of the original RNA sample between 85–160 nt (Figure 2B, left gel; labelled by a blue bar). Also excise the distinct band of the oGAB11 control (Figure 2B, right gel; band is labelled by a yellow asterisk). Transfer the gel slices to two pierced (20G needle) 0.5 ml RNase/DNase-free tubes. Put each 0.5 ml RNase/DNase-free tube into a 1.5 ml RNase/DNase-free microcentrifuge tubes. Centrifuge for 3 min at 20,000 g at room temperature.
The excision of the dephosporylated oGAB11 control is optional. -
54
Add 200 μl RNase/DNase-free H2O to each tube and incubate for 10 min at 70°C. Vortex for 30 sec and transfer the gel slurry to microcentrifuge tube filters. Centrifuge for 3 min at 20,000 g at room temperature.
-
55
Transfer the eluate to new 1.5 ml RNase/DNase-free non-stick tubes. Add 2 μl GlycoBlue (15 mg/ml), 15 μl 5 M NaCl and mix.
-
56
Add 750 μl isopropanol and mix. Precipitate cDNA for ≥ 1 h at −20°C. Centrifuge for 30 min at 20,000 g at 4°C. Remove supernatant and wash the cDNA pellet with 750 μl pre-cooled 80% (vol/vol) ethanol.
-
57
Air-dry the cDNA pellet for 10 min and resuspend pellet in 15 μl 10 mM Tris-HCl (pH 8.0).
cDNA circularization & PCR amplification
-
58
Prepare the circularization mix for the cDNA sample and the oGAB11 control as follows:
Component Amount per reaction (μl) Final CircLigase 10x Reaction Buffer 2.0 1x ATP (1 mM) 1.0 50 μM MnCl2 (50 mM) 1.0 2.5 mM -
59
Add 4 μl of circularization mix to 15 μl cDNA sample and the oGAB11 control (step 57) and mix. Add 1 μl (100 U) of CircLigase and mix.
-
60
Incubate 60 min at 60°C followed by 10 min at 80°C.
The circularized cDNA can be stored at −20°C up to one year, if not proceeded immediately. -
61
Prepare PCR master mixes for 4 test reactions for the cDNA sample and for the oGAB11 control as follows:
Component Amount for 4.5 reactions (μl) Final Phusion HF Buffer, 5x 18 1x dNTPs (10 mM) 1.8 0.2 mM Forward primer (Illumina Index primer, 100 μM) 0.45 0.5 μM oNTI231 (Reverse primer, 100 μM) 0.45 0.5 μM DNase-free H2O 63.9 Phusion DNA Polymerase (2 U/μl) 0.9 1.8 U The oGAB11 control serves as a positive control for the PCR amplification. -
62
For each reaction place 19 μl PCR master mix into a 0.2 ml PCR tube. Add 1.0 μl circularized cDNA of the original RNA sample or of the oGAB11 control (step 60) and mix.
-
63
Perform PCR test amplifications as follows:
Cycle number Denature Anneal Extend 1 98°C, 30 sec 2–14 98°C, 10 sec 60°C, 10 sec 72°C, 5 sec Stop the PCR amplification by removing the PCR tube at the end of the extension step, typically after 6, 8, 10 and 12 rounds of amplification. -
64
Add 3.4 μl 6x DNA loading dye to each completed PCR reaction and mix. Prepare DNA control ladder. Add 1.0 μl DNA control ladder to 9 μl DNase-free H2O. Add 2 μl 6x DNA loading dye and mix.
-
65
Load the PCR reactions and DNA control ladder on an 8% (wt/vol) TBE gel, and run at 180 V for 45 min.
-
66
Stain the gel in 50 ml Gel staining solution for 5 min at room temperature. Visualize the gel and identify the optimal PCR amplification cycle for the cDNA sequencing library.
At the optimal amplification cycle a clear band at ~120 nt is visible, whereas a DNA smear that migrates slower in an 8% TBE-gel is absent. -
67
Perform 4 PCR reactions per sample (not for the oGAB11 control) with the adjusted amplification cycle as described in steps 61 to 66.
-
68
Excise the PCR product from the gel and transfer the gel slice into a pierced (20G needle) 0.5 ml RNase/DNase-free tube. Put the 0.5 ml RNase/DNase-free tube into a 1.5 ml RNase/DNase-free microcentrifuge tube. Centrifuge for 3 min at 20,000 g at room temperature.
-
69
Add 670 μl DNA soaking buffer and mix. Incubate at room temperature over-night at 500 g.
-
70
Transfer the gel slurry to a Microfuge tube filter and centrifuge for 3 min at 20,000 g at room temperature.
-
71
Transfer the eluate to a new 1.5 ml RNase/DNase-free non-stick tube. Add 2 μl GlycoBlue (15 mg/ml), 680 μl isopropanol and mix. Precipitate the subRNA-seq library for ≥ 60 min at −20°C.
-
72
Centrifuge for 30 min at 20,000 g at 4°C. Discard the supernatant and wash the pellet with 750 μl 80% (vol/vol) ethanol. Air-dry pellet.
-
73
Resuspend the subRNA-seq library in 10 μl 10 mM Tris-HCl (pH 8.0).
The subRNA-seq library can be stored at −20°C up to one year, if not proceeded immediately. -
74
The quantity and quality of the subRNA-seq library is determined by using the Qubit dsDNA HS Assay Kit and the Agilent Bioanalyzer High Sensitivity DNA Analysis Kit according to the manufacturer’s instructions.
The expected concentration is 30 to 70 nM. The subRNA-seq library is ready for high-throughput sequencing on an Illumina sequencing platform using the custom sequencing primer oLSC006 (Table 1).
REAGENTS AND SOLUTIONS
Use RNase- and DNase-free H2O in all recipes and experimental protocol steps.
Cytoplasmic lysis buffer
0.15% (vol/vol) NP-40
10 mM Tris-HCl (pH 7.0)
150 mM NaCl
25 μM α-amanitin
10 U SUPERase.IN
1x Protease inhibitor mix
Prepare this buffer freshly before use and store on ice.
Sucrose buffer
10 mM Tris-HCl (pH 7.0)
150 mM NaCl
25% (wt/vol) sucrose
25 μM α-amanitin
20 U SUPERase.IN
1x Protease inhibitor mix
Prepare this buffer freshly before use and store on ice.
Nuclei wash buffer
0.1% (vol/vol) Triton X-100
1mM EDTA
25 μM α-amanitin
40 U SUPERase.IN
1x Protease inhibitor mix
In 1x PBS
Prepare this buffer freshly before use and store on ice.
Glycerol buffer
20 mM Tris-HCl (pH 8.0)
75 mM NaCl
0.5 mM EDTA
50% (vol/vol) glycerol
0.85 mM DTT
25 μM α-amanitin
10 U SUPERase.IN
1x Protease inhibitor mix
Prepare this buffer freshly before use and store on ice.
Nuclei lysis buffer
1% (vol/vol) NP-40
20 mM HEPES (pH 7.5)
300 mM NaCl
1M urea
0.2 mM EDTA
1mM DTT
25 μM α-amanitin
10 U SUPERase.IN
1x Protease inhibitor mix
Prepare this buffer freshly before use and store on ice.
2x Alkaline fragmentation solution
100 mM NaCO3 (pH 9.2)
For 5 ml: mix 0.6 ml of 0.1 M Na2CO3 and 4.4 ml of 0.1 M NaHCO3
Store in air-tight screw-cap tubes at room temperature for up to 4 months.
RNA precipitation solution
For one reaction (562 μl): mix 60 μl of 3 M sodium acetate (pH 5.5), 2 μl of GlycoBlue (15 mg/ml) and 500 μl of RNase/DNase-free H2O.
Prepare this solution freshly before use and store on ice.
Gel staining solution
For 50 ml (enough for one gel): add 5 μl of SYBR Gold nucleic acid gel stain to 50 ml of 1x TBE.
Prepare this solution freshly before use and store protected from light at room temperature.
SUPERase.IN/DTT mix
For one reaction (1.3 μl): mix 0.5 μl of SUPERase.IN (20 U/μl) and 0.8 μl of 0.1 M DTT.
Prepare this solution freshly before use and store on ice.
DNA soaking buffer
For one reaction (668 μl): mix 6.7 μl of 1 M Tris-HCl (pH 8.0), 40 μl of 5 M NaCl, 1.3 μl of 0.5 M EDTA and 620 μl of RNase/DNase-free H2O.
Prepare this solution freshly before use and store at room temperature.
COMMENTARY
Background Information
High-throughput sequencing of subcellular RNAs has been used by us and others to study nascent transcription, splicing, RNA processing, RNA transport and decay (Mayer et al., 2015; Pandya-Jones and Black, 2009; Bhatt et al., 2012; Khodor et al., 2011; Ferrari et al., 2013; Tilgner et al., 2012; Zaghlool et al., 2013; Conrad et al., 2014; Mondal et al., 2010; Weber et al., 2014). Most of these studies have focused on specific subsets of RNAs such as chromatin-associated RNAs (Khodor et al., 2011; Mondal et al., 2010). However, a detailed experimental protocol for the analysis of the identity and abundance of the full spectrum of Pol II-synthesized RNAs in different cellular compartments, ranging from unstable chromatin-associated nascent RNAs to fully-processed steady-state cytoplasmic RNAs, is lacking.
Here, we provide step-by-step guidance to interrogate RNAs in the chromatin, nucleoplasmic and cytoplasmic fractions of mammalian cells. The subRNA-seq approach combines effective cell fractionation with efficient and robust sequencing library preparation, thus providing a snapshot of the subcellular distribution of RNAs at the time when cells are lysed.
The cell fractionation is based on protocols originally developed in the Schibler, Black and Smale laboratories (Wuarin and Schibler, 1994; Pandya-Jones and Black, 2009; Bhatt et al., 2012), and was optimized by us to minimize cross-contamination of RNAs between the different cellular compartments and to avoid run-off transcription during sample processing (Mayer et al., 2015). Cells are lysed by using the mild detergent NP-40 which keeps the nuclear membrane intact. The released nuclei are separated from the cytoplasmic fraction by centrifugation through a sucrose cushion. The nucleoplasm is separated from the chromatin in the presence of urea, NP-40 and NaCl. This fast and effective chromatin fractionation method exploits the high stability of the Pol II transcription elongation complex, even in the presence of high salt, detergents and urea (Cai and Luse, 1987; Wuarin and Schibler, 1994). In contrast to ionic detergents, such as sarkosyl, urea removes most of the chromatin-associated proteins except for transcribing RNA polymerase and histone proteins (Wuarin and Schibler, 1994; Khodor et al., 2011). The chromatin is kept in a compacted state that can be collected by low-speed centrifugation. No ultracentrifugation is required.
The DNase-treated subcellular RNAs are then applied to the sequencing library preparation. The method described here is based on protocols originally developed for NET-seq (Churchman and Weissman, 2012a; Mayer et al., 2015; Churchman and Weissman, 2012b; Mayer and Churchman, 2016). In contrast to the NET-seq approach, which relies on high-throughput sequencing of the 3′-ends of nascent transcripts, subRNA-seq sequences across entire transcripts. The subRNA-seq library generation differs in two important ways from the original NET-seq protocol:
RNA fragmentation is performed before DNA linker-ligation.
The fragmented RNAs are dephosphorylated to generate an accessible 3′-OH prior to the DNA linker-ligation.
This library preparation method results in DNA strand-specific sequencing libraries for the cytoplasmic, nucleoplasmic and chromatin-associated RNAs and are ready for deep sequencing on Illumina platforms using a custom sequencing primer.
The subRNA-seq approach has been successfully used for the analysis of subcellular transcriptomes in HeLa S3 and HEK293T cells (Mayer et al., 2015). In the meantime, the authors have successfully applied the cell fractionation approach to several other mammalian cell types including K562, MOLT4, primary human foreskin fibroblasts (Padovan-Merhar et al., 2015) and murine NIH/3T3 cells. Since the cell fractionation is most critical for the success of a subRNA-seq experiment, the authors expect that the protocol described here should also be applicable to any other mammalian cell type.
Critical Parameters & Troubleshooting
subRNA-seq is a quantitative genome-wide approach. This requires that each key step of the protocol works as efficient as possible. The following steps of subRNA-seq are most critical and their success needs to be monitored.
Cell fractionation
Cell fractionation is the most critical step of the subRNA-seq protocol and needs to be as efficient as possible in order to avoid cross-contamination between the subcellular fractions. Although the cell fractionation protocol was extensively optimized (Mayer et al., 2015; Mayer and Churchman, 2016), we recommend to monitor its success by Western blotting using antibodies raised against subcellular marker proteins such as GAPDH (cytoplasmic marker), U1 snRNP70 (nucleoplasmic marker), Histone 2B (chromatin marker) and transcribing RNA polymerase II (Pol II Ser2-P, chromatin marker) (Figure 2A).
When cytoplasmic marker proteins are detected in the chromatin fraction, we recommend to reduce the amount of cells per fractionation. Using more than 2x 107 cells per fractionation can reduce the cell lysis efficiency and thus disrupt effective fractionation. Furthermore, we recommend to completely remove the supernatant after washing the cell nuclei (protocol 1, step 11).
Strong Western blot signals of transcribing Pol II (Pol II Ser2-P) in the cytoplasmic fraction indicate that the treatment during cell lysis is too harsh leading to dissociation of elongating Pol II from the chromatin. In this case, we recommend to carefully reduce the amount of cell lysis buffer that is added to the cell pellet (protocol 1, step 5).
Optionally, quantitative real-time PCR can be performed to assess whether intron-containing nascent RNAs are enriched in the chromatin fraction. This can be accomplished by using primer pairs directed against selected intronic and exonic regions of well-expressed transcripts in the respective cell-type of interest. Intronic containing RNAs should be enriched in the chromatin fraction whereas intron-less fully-processed RNAs should be prevalent in the cytoplasmic fraction (Figure 2C).
3′ RNA dephosphorylation
Only RNA molecules with a 3′-OH will ligate with the DNA linker and are captured in the sequencing library. In order to avoid biases and to make subRNA-seq a quantitative approach, the removal of 3′-phosphates should occur on ≥ 90% of the RNAs. Therefore, we recommend to monitor the success of 3′-dephosphorylation by using a 3′-phosphorylated RNA control oligo (oGAB11-P, Table 1 and Figure 2B, right panel). In case of a low dephosphorylation efficiency, we recommend the following protocol adjustments: (i) increase the concentration of the T4 PNK, (ii) use a new batch of T4 PNK (different lot number) (iii) increase the T4 PNK treatment time, (iv) decrease the amount of RNA per dephosphorylation reaction.
DNA linker ligation
Only RNA molecules that ligate to the DNA linker are captured in the final subRNA-seq and therefore the ligation should be as efficient as possible (≥90%). Only in this case the final subRNA-seq library quantitatively reflects the pool of RNAs in the original sample. The ligation efficiency should be tested by using the RNA control oligo oGAB11 (Table 1 and Figure 2). We noticed that the ligation efficiency can vary with different batches of the truncated T4 RNA Ligase 2. We recommend to keep track of the lot numbers of the ligase. Whenever the ligation efficiency is below 90%, we recommend to use a new batch of enzyme (different lot number). Furthermore, increasing the ligase concentration and the ligation time can enhance the ligation efficiency. We also recommend to replace components of the ligation reaction every 4 months.
Anticipated Results
The RNA yield from the cytoplasmic, nucleoplasmic and chromatin fractions obtained from 1x 107 HeLa S3 cells is ~150 μg, ~35 μg and ~25 μg, respectively. The A260/A280 ratio is ~2.1 as determined by NanoDrop spectrophotometer measurement. The efficiency of the 3′-dephosphorylation of fragmented RNA is typically ≥90% as monitored by PAGE loading the cDNA obtained from the oGAB11-P and the oGAB11 RNA control oligos (Table 1 and Figure 2B, right panel). The efficiency of the DNA linker ligation is usually ≥ 95% as determined by PAGE using the RNA control oligo oGAB11 (Table 1 and Figure 2, right panel). The total yield of a typical subRNA-seq library is 20–30 ng, as determined by Qubit fluorometer and Bioanalyzer measurements. The average fragment length of a subRNA-seq library is ~150 nt. The RNA from the chromatin fraction is enriched for intron-containing nascent RNAs as shown by a lower exon/intron ratio as compared to RNAs from the nucleoplasmic and cytoplasmic fractions (Figure 2C).
Time Considerations
On day 1, cell fractionation (protocol 1, steps 1–16), RNA isolation (protocol 1, steps 17–25), DNase I digest (protocol 1, steps 26 and 27) and the rRNA depletion (protocol 2, optional steps 1–7) is performed what takes approximately 6 hours. On day 2, the RNA is fragmented (protocol 2, steps 8–27) and dephosphorylated (protocol 2, steps 28–35). This will take approximately 5 to 6 hours. On day 3, the DNA linker is ligated (protocol 2, steps 36–42) and the cDNA synthesis (protocol 2, steps 43–57) is conducted. This requires ~5 to 6 hours. On day 4, the cDNA is circularized and the subRNA-seq libraries are amplified by a limited number of PCR cycles (protocol 2, steps 58–69). This will take ~6 hours and requires an overnight elution. On day 5, subRNA-seq library preparation is completed and the quality is monitored (protocol 2, steps 70–74). This will require ~2 to 3 hours. If the libraries pass the quality control, they are ready for sequencing on the Illumina sequencing platform.
The protocol includes many pausing points and thus can be performed in a flexible manner and according to the working habits of the experimenter.
Acknowledgments
We thank Mirjam Arnold for critical comments on the manuscript. We thank Julia di Iulio for help in analyzing subRNA-seq data. This work was supported by US National Institutes of Health National Human Genome Research Institute (NHGRI) grant R01HG007173 to L.S.C.; a Damon Runyon Dale F. Frey Award for Breakthrough Scientists (to L.S.C.); and a Burroughs Wellcome Fund Career Award at the Scientific Interface (to L.S.C.). A.M. was supported by Long-Term Postdoctoral Fellowships of the Human Frontier Science Program (HFSP) (LT000314/2013-L) and the European Molecular Biology Organization (EMBO) (ALTF858-2012).
LITERATURE CITED
- Brueckner F, Cramer P. Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nature Structural & Molecular Biology. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- Cai H, Luse DS. Transcription initiation by RNA polymerase II in vitro. The Journal of Biological Chemistry. 1987;262:298–304. [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2012a;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Native Elongating Transcript Sequencing (NET-seq) Current Protocols in Molecular Biology. 2012b:4.14.1–4.14.17. doi: 10.1002/0471142727.mb0414s98. [DOI] [PubMed] [Google Scholar]
- Conrad T, Marsico A, Gehre M, Ørom UA. Microprocessor Activity Controls Differential miRNA Biogenesis In Vivo. Cell Reports. 2014;9:542–554. doi: 10.1016/j.celrep.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2013;488:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F, Plachetka A, Alekseyenko AA, Jung YL, Ozsolak F, Kharchenko PV, Park PJ, Kuroda MI. “‘Jump Start and Gain’” Model for Dosage Compensation in Drosophila Based on Direct Sequencing of Nascent Transcripts. Cell Reports. 2013;5:629–636. doi: 10.1016/j.celrep.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Genome-wide annotation and quantitation of translation by ribosome profiling. Current protocols in molecular biology. 2013;Chapter 4(Unit 4.18) doi: 10.1002/0471142727.mb0418s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature Protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D. Dealing with Pervasive Transcription. Mol Cell. 2013;52:473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CHA, Marr MT, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes & Development. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17:227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Mayer A, Churchman LS. Genome-wide profiling of RNA polymerase transcription at nucleotide resolution in human cells with native elongating transcript sequencing. Nature Protocols. 2016;11:813–833. doi: 10.1038/nprot.2016.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native Elongating Transcript Sequencing Reveals Human Transcriptional Activity at Nucleotide Resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinel DM, Sträßer K. Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae. BioEssays. 2015;37:666–677. doi: 10.1002/bies.201400220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Research. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D. Biochimica et Biophysica Acta. BBA - Gene Regulatory Mechanisms. 2012;1819:494–506. doi: 10.1016/j.bbagrm.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, Foley SW, Wu AR, Churchman LS, Singh A, Raj A. Single Mammalian Cells Compensate for Differences in Cellular Volume and DNA Copy Number through Independent Global Transcriptional Mechanisms. Mol Cell. 2015:1–15. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript Dynamics of Proinflammatory Genes Revealed by Sequence Analysis of Subcellular RNA Fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Research. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015:1–12. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes Are Context-Specific, H2A.Z-Modulated Barriers to RNA Polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Molecular and Cellular Biology. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool A, Ameur A, Nyberg L, Halvardson J, Grabherr M, Cavelier L, Feuk L. Efficient cellular fractionation improves RNA sequencing analysis of mature and nascent transcripts from human tissues. BMC Biotechnology. 2013;13:99. doi: 10.1186/1472-6750-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]