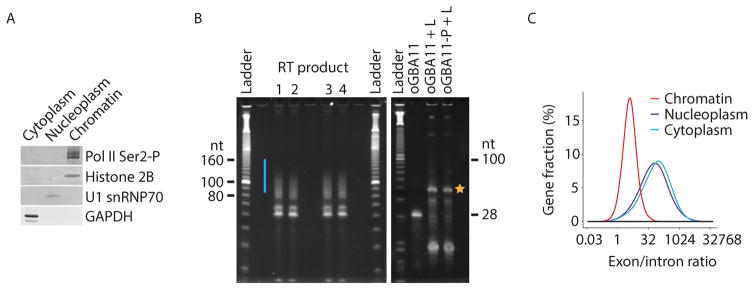
Figure 2. Key quality controls and expected results from subRNA-seq experiments.
(A) Representative Western blot of subcellular fractions. The different cellular fractions obtained from HeLa S3 cells were probed against transcribing Pol II (Pol II Ser2-P; 3E8 antibody) the chromatin marker Histone 2B (FL-126 antibody), the nucleoplasmic marker U1 snRNP70 (C-18 antibody) and the cytoplasmic marker GAPDH (6C5 antibody). The volumes of the different cellular fractions have been adjusted so that the Western blot signals can be compared between the fractions. This figure was modified from (Mayer et al., 2015).
(B) Representative gels showing the product of the reverse transcription (RT) of the samples (left panel) and of the oGAB11 controls (right panel). The region that is excised from the gel is indicated by a blue bar or yellow star for the samples and the oGAB11 controls, respectively.
(C) Distributions of the size-normalized ratio of subRNA-seq reads that map to exons versus introns for each gene. subRNA-seq libraries were generated from HeLa S3 cells. This figure was adapted from (Mayer et al., 2015).