Figure 2.
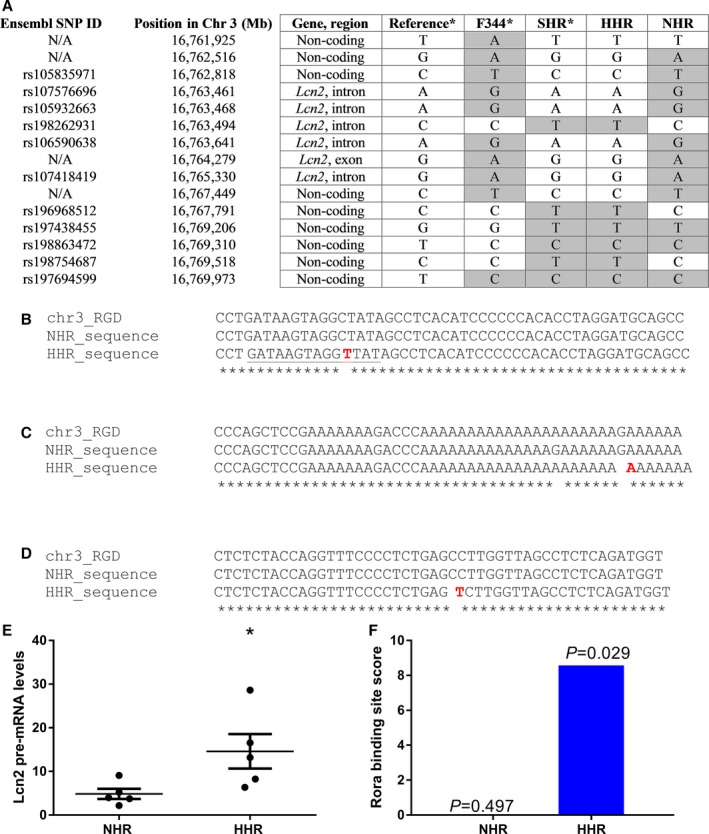
Variants in the lipocalin‐2 (Lcn2) gene, showing regions with variants in the HHR, according to the Rat Genome Database (RGD; version 5). A, Genotype analysis of the region of 10 000 bp around the Lcn2 gene, showing the origin of the variants observed in the HHR. Highlighted in gray are variants that differ from the reference genome, showing that the HHR carries 3 unique variants which were inherited from the SHR. B, Single‐nucleotide polymorphism (SNP) on chr3: 16 767 791 (rs196968512 C/T) 1401 bp upstream of Lcn2 gene. C, SNP on chr3: 16 767 398 (G/A) in a highly conserved region 1001 bp upstream the Lcn2 gene. D, Nonfunctional intronic SNP originally from SHR on position chr3: 16 763 494 (rs198262931 C/T). E, Lcn2 pre‐mRNA is also upregulated in the HHR compared to the NHR (n=5/strain), suggesting that it is dysregulated at the transcriptional level. F, The SNP, rs196968512, creates a new binding site for the transcription factor, Rora (region underlined in B), which acts as an enhancer for expression of Lcn2 and is exclusive of the HHR. The figure shows the binding site score and the P values for the binding of Rora to that region. chr3 indicates rat chromosome 3; F344, Fisher 344 rat; HHR, hypertrophic heart rat; N/A, nonannotated SNP; NHR, normal heart rat; RGD, reference sequence from the Rat Genome Database v5; SHR, spontaneously hypertensive rat.
