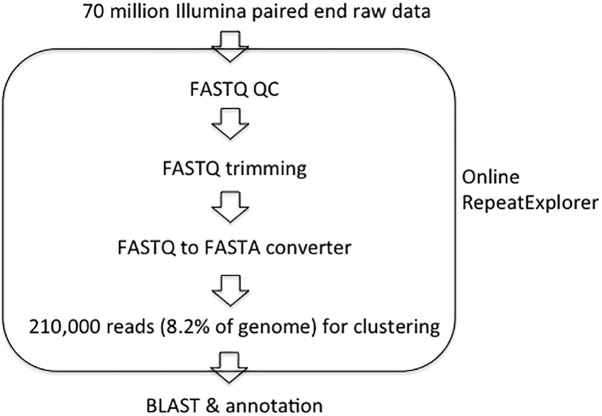
Figure 1. Workflow for repeat analysis in this study.
Raw data from next-generation sequencing were uploaded to the Galaxy-based RepeatExplorer platform. The FASTQ QC: READ QC tool was then used to verify the quality of the reads before removing unnecessary sequences (i.e., adapter sequences) from the ends of each read using the FASTQ Trimmer tool. The QC analysis was then repeated, and the FASTQ to FASTA converter tool was used to convert each read into FASTA format. Using these DNA sequence reads as input, sequences undergo clustering, during which an “all-to-all” sequence comparison is performed, and similar sequences are grouped together into clusters.