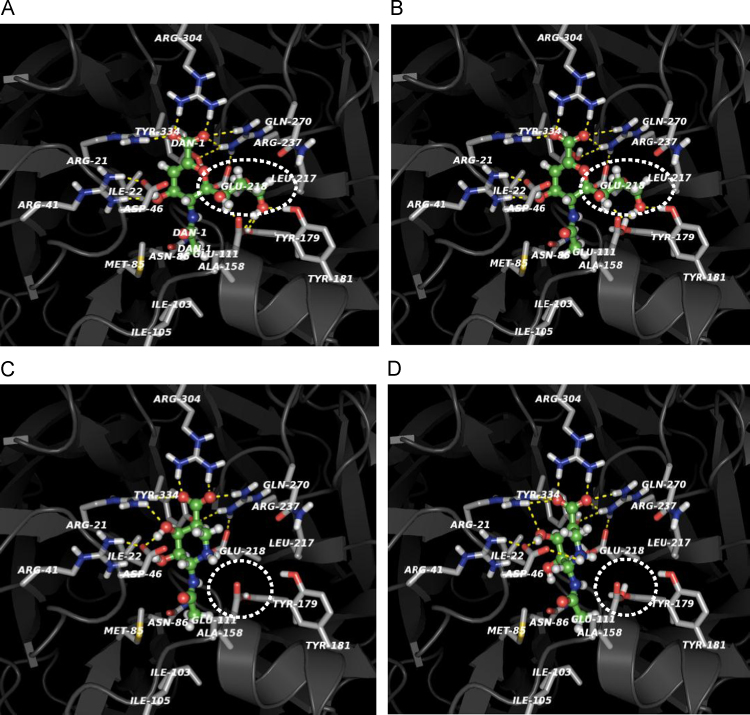
Fig. 4.
Molecular docking of the DANA and siastatin B to the active pocket of NEU2 based on the crystal structure of the NEU2/DANA complex (PDB code: 1VCU) without steric hindrance. Protonation of amino acid residues is indicated around inhibitor binding site compared at pH 4.0 and pH 6.0. Comparison of binding manners of DANA at pH 4.0 (A) and pH 6.0 (B). pH-dependent neutralization of E111 is predicted to have influence on binding of DANA C7–9 fragment (dashed oval). The E218 interacts with R237 via ionic bond. Neutralization of E218 may possibly have the effect on local structure of NEU2 supporting the binding of DANA C7–9 fragment. Comparison of binding manners of siastatin B at pH 4.0 (C) and pH 6.0 (D). There is no chemical groups in siastatin B corresponding to those of DANA C7–9 fragment, which may not influence the interaction between siastatin B and the neutralized amino acid residues (E111 and E218, dashed circle) at pH 4.0. The imino group (1NH) can interact with D46, which is not influenced by neutralization at pH 4.0.