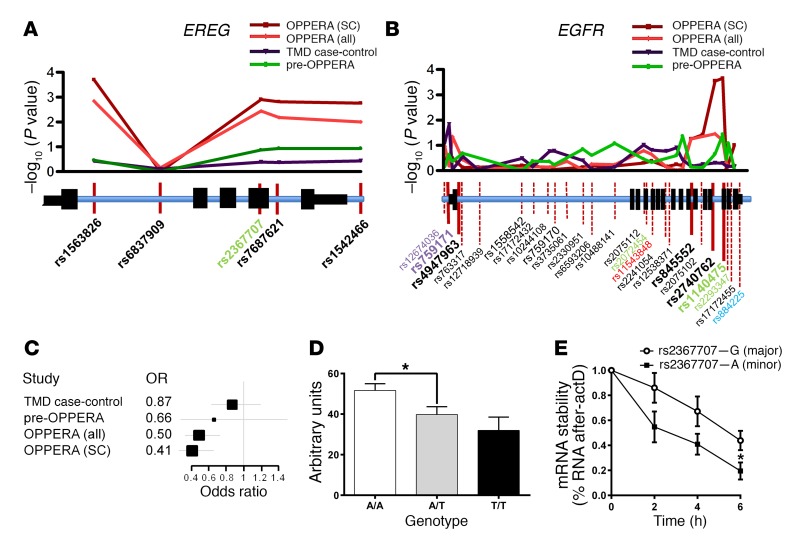
Figure 3. Genetic association of EREG and EGFR with TMD pain.
Genetic association of EREG (A) and EGFR (B) SNPs with chronic TMD pain in OPPERA cases vs. super controls (SC), OPPERA cases vs. controls (all), TMD case-control cohort, and pre-OPPERA cohorts (see Methods). Manhattan plots and corresponding gene loci are shown where position of tested SNPs is given relative to gene structure. Nonsynonymous (red), synonymous (green), promoter region (gray), and 3′ intragenic region (blue) SNPs are indicated. The pattern of association within the EREG gene locus was identical in all cohorts. The pattern of association for the EGFR gene locus revealed some differences between cohorts; however, the 5′ and 3′ ends of the gene consistently showed association with elevated TMD risk. The EGFR SNPs showing association in independent SNP tests (marked in bold) were used for haplotype analysis (see Supplemental Table 5). (C) Forest plot depicting odds ratios (OR; with 95% confidence intervals) for minor allele T of rs1563826 in 4 human chronic pain cohorts. (D) Association of EREG mRNA level with EREG rs1563826 in the TMD case-control cohort measured by quantitative RT‑PCR. Bars represent mean ± SEM for EREG expression in leukocytes expressed in arbitrary units relative to GAPDH (see Methods). One-way ANOVA revealed a significant difference among genotypes; F2,247 = 3.7, P = 0.03. *P < 0.05; the A/A versus T/T comparison was P = 0.053, likely due to the small number of T/T homozygotes. (E) Following actD treatment, the rate of mRNA degradation was significantly lower for cells expressing WT (G allele) EREG mRNA compared with those the minor A allele of rs2367707 (2-tailed paired t test, t4 = 2.8, P = 0.05). Symbols represent mean ± SEM for percentage of mRNA expression compared with time 0; n = 3/genotype.