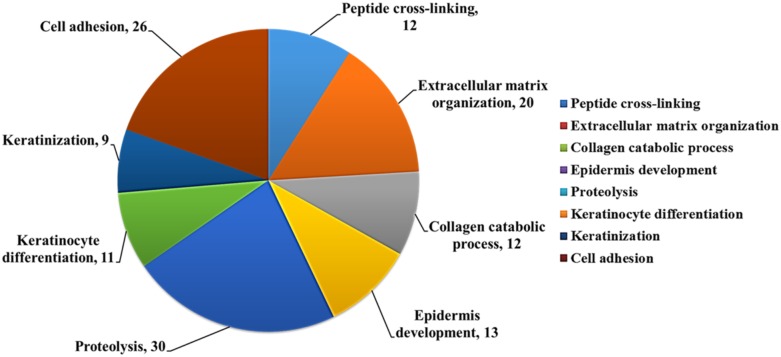
Figure 5. Biological function analysis of differentially expressed genes in asthmatic bronchial epithelial cells.
Functional analysis of 504 differentially expressed genes discovered from the GE microarray data was performed by functional annotation (Biological Processes) in DAVID database. The results showed that these genes were involved in cell adhesion (26 genes), keratinization (9 genes), keratinocyte differentiation (11 genes), proteolysis (30 genes), collagen catabolic process (12 genes), extracellular matrix organization (20 genes), and peptide cross-linking (12 genes). The selected criteria were EASE = 0.1, p-value < 0.05, and fold enrichment > 1.3.