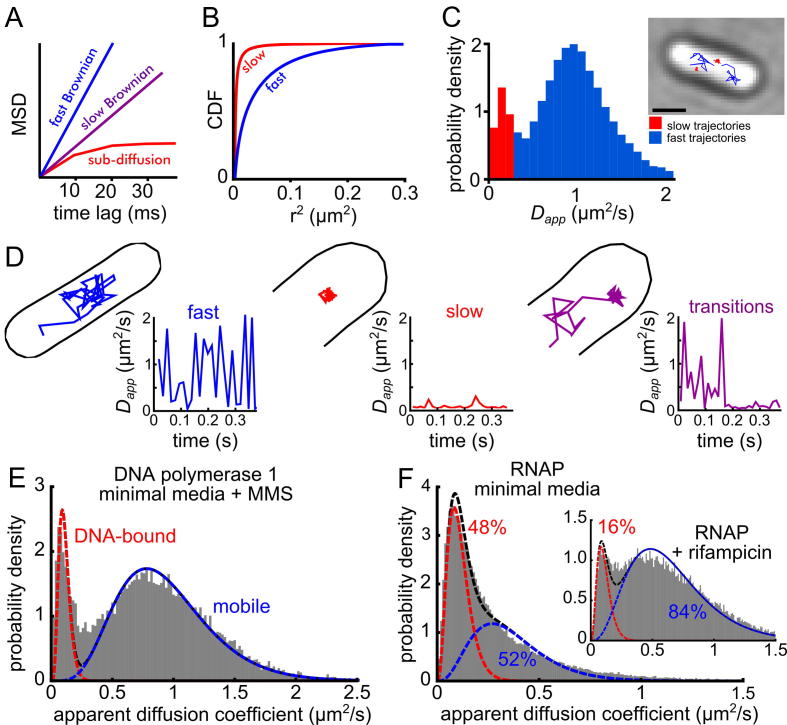
Fig. 4.
Analyzing single-particle tracking PALM data. A) Plotting the mean squared displacement of the sptPALM trajectories against time lag can provide information about the mobility of the labelled protein, and establish if motion is Brownian (where MSD increases linearly with increasing time lag) or sub-diffusive. B) Cumulative distribution of the squared displacements. This distribution can be fitted with Eqs. (2), (3), to extract information about the mobility of the proteins and the number of diffusive species. C) Distribution of apparent diffusion coefficients (Dapp) calculated for each single-molecule trajectory. A threshold can be used to sort individual trajectories based on their Dapp value, as shown in the example cell with slow trajectories colored red and fast trajectories colored blue (right). D) Examples of long trajectories (ten or more localizations) classified according to their Dapp transitions: a fast diffusing molecule, with a high average Dapp value over the whole trajectory (blue), a slow-moving molecule, with a low average Dapp value (red), and a molecule undergoing transition from fast (high Dapp) to slow (low Dapp) (purple). E) The Dapp distribution for DNA polymerase 1 treated with a DNA damaging agent to recruit molecules to DNA. The distribution shows two clearly resolvable peaks, which can be fitted with a two-species model (using Eq. (6)) to extract fractions of molecules in the low-mobility DNA-bound state, and the mobile state. F) The distribution of RNAP Dapp values can also be fitted with a two-species model. Treatment with rifampicin blocks transcription, causing a large drop in the fraction of DNA-bound RNAPs (inset). Panels A–C adapted from [73]. Panel D adapted from Ref. [55]. Panels E,F adapted from [65]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)