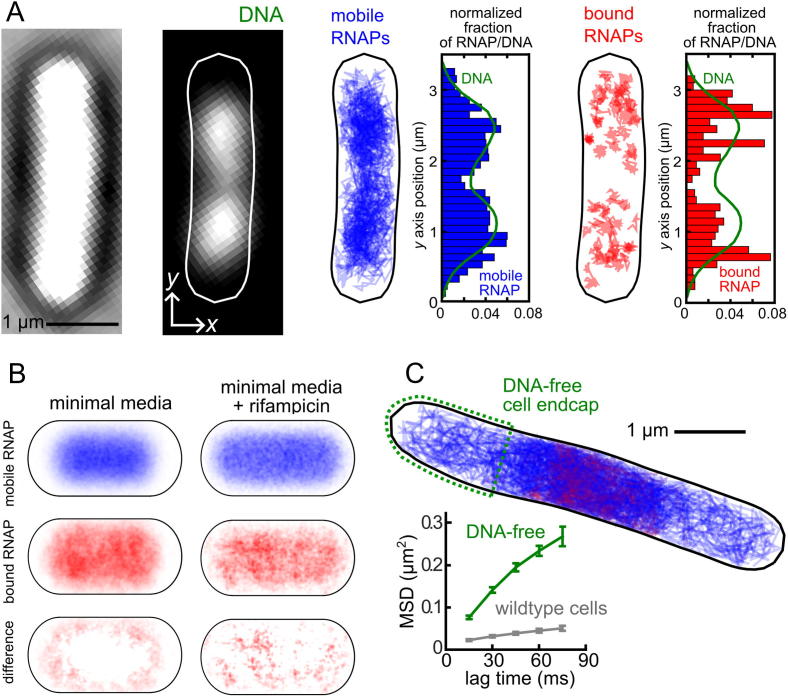
Fig. 5.
Spatial organization of transcription and non-specific DNA interactions. A) Transmission image of an example cell, and an image of DNA stained with an intercalating fluorescent dye. The distribution of sorted mobile RNAP trajectories (blue lines/bars) closely matched the distribution of DNA (green line). The distribution of bound RNAPs in the same example cell shows a more clustered distribution which does not closely follow the distribution of DNA. B) Spatial distribution of sorted RNAP trajectories averaged over ∼200 cells between 1.6 and 2.5 µm long. Transcribing RNAPs show a bias towards the periphery of the nucleoid region, which is lost after blocking elongating RNAPs with rifampicin. C) Example ‘minimal-DNA’ cell (top); temperature-sensitive DnaC mutant cells are grown at a non-permissive temperature to give long cells with a single centrally located chromosome. Tracking RNAPs only in the DNA-free cell endcaps (green dashed region) allows the free 3D diffusion to be determined. The mean squared displacement (bottom) shows that the diffusion of RNAP in DNA-free cell endcaps (green line) is much faster than the average diffusion of RNAP molecules in normal unperturbed cells (grey line). Panels A–C adapted from [65]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)