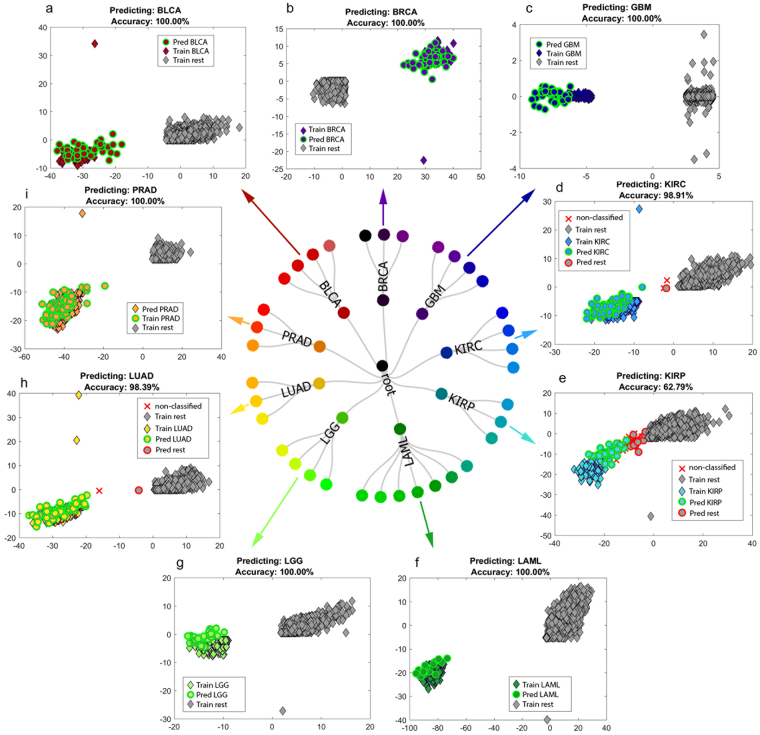
Figure 4.
Representative leave-one-subtype-out scores plots. Discrimination plots generated for the prediction of: (a) ‘squamous’ subtype to bladder urothelial carcinoma (BLCA); (b) ‘reactive-like’ subtype to breast adenocarcinoma (BRCA); (c) ‘classical’ subtype to glioblastoma multiforme (GBM); (d) ‘ccB(2)’ subtype to kidney renal clear cell carcinoma (KIRC); (e) ‘PRCC Type 2’ subtype to kidney renal papillary cell carcinoma (KIRP); (f) ‘FAB M5’ subtype to acute myeloid leukemia (LAML); (g) ‘IDHmut-codel’ to lower grade glioma (LGG); (h) ‘proximal proliferative’ to lung adenocarcinoma (LUAD); and (i) ‘ETS-fusion negative’ to prostate adenocarcinoma (PRAD). Correctly predicted samples are indicated by green outline, a red ‘x’ denotes non-classified samples and misclassified samples are indicated by red outline. Axes represent discriminant components, where the second component is used only for visualization purposes. The scores plotted are obtained from the ‘best’-chosen dimensionality reduction space. Subtype assignment information is provided in the Methods and Supplementary Information Note 1. Most subtypes were correctly assigned to the respective cancer type with 95–100% accuracy (see Supplementary Table 4).